Guanidinium chloride denaturation of the dimeric Bacillus licheniformis BlaI repressor highlights an independent domain unfolding pathway
- PMID: 15285720
- PMCID: PMC1134101
- DOI: 10.1042/BJ20040658
Guanidinium chloride denaturation of the dimeric Bacillus licheniformis BlaI repressor highlights an independent domain unfolding pathway
Abstract
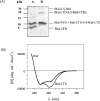
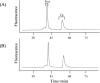
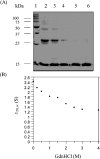
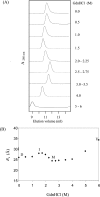
The Bacillus licheniformis 749/I BlaI repressor is a prokaryotic regulator that, in the absence of a beta-lactam antibiotic, prevents the transcription of the blaP gene, which encodes the BlaP beta-lactamase. The BlaI repressor is composed of two structural domains. The 82-residue NTD (N-terminal domain) is a DNA-binding domain, and the CTD (C-terminal domain) containing the next 46 residues is a dimerization domain. Recent studies have shown the existence of the monomeric, dimeric and tetrameric forms of BlaI in solution. In the present study, we analyse the equilibrium unfolding of BlaI in the presence of GdmCl (guanidinium chloride) using different techniques: intrinsic and ANS (8-anilinonaphthalene-l-sulphonic acid) fluorescence, far- and near-UV CD spectroscopy, cross-linking, analytical ultracentrifugation, size exclusion chromatography and NMR spectroscopy. In addition, the intact NTD and CTD were purified after proteolysis of BlaI by papain, and their unfolding by GdmCl was also studied. GdmCl-induced equilibrium unfolding was shown to be fully reversible for BlaI and for the two isolated fragments. The results demonstrate that the NTD and CTD of BlaI fold/unfold independently in a four-step process, with no significant co-operative interactions between them. During the first step, the unfolding of the BlaI CTD occurs, followed in the second step by the formation of an 'ANS-bound' intermediate state. Cross-linking and analytical ultracentrifugation experiments suggest that the dissociation of the dimer into two partially unfolded monomers takes place in the third step. Finally, the unfolding of the BlaI NTD occurs at a GdmCl concentration of approx. 4 M. In summary, it is shown that the BlaI CTD is structured, more flexible and less stable than the NTD upon GdmCl denaturation. These results contribute to the characterization of the BlaI dimerization domain (i.e. CTD) involved in the induction process.
Figures







Similar articles
-
Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.Nucleic Acids Res. 2007;35(13):4384-95. doi: 10.1093/nar/gkm448. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576674 Free PMC article.
-
The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP beta-lactamase.Mol Microbiol. 2002 May;44(3):685-94. doi: 10.1046/j.1365-2958.2002.02888.x. Mol Microbiol. 2002. PMID: 12022149
-
The C-terminal domain of dimeric serine hydroxymethyltransferase plays a key role in stabilization of the quaternary structure and cooperative unfolding of protein: domain swapping studies with enzymes having high sequence identity.Protein Sci. 2004 Aug;13(8):2184-95. doi: 10.1110/ps.04769004. Protein Sci. 2004. PMID: 15273312 Free PMC article.
-
Urea and thermal equilibrium denaturation studies on the dimerization domain of Escherichia coli Trp repressor.Biochemistry. 1997 May 13;36(19):5612-23. doi: 10.1021/bi970056e. Biochemistry. 1997. PMID: 9153401
-
Characterization of an unfolding intermediate and kinetic analysis of guanidine hydrochloride-induced denaturation of the colicin E1 channel peptide.Biochemistry. 1997 Mar 11;36(10):3037-46. doi: 10.1021/bi961926f. Biochemistry. 1997. PMID: 9062135
Cited by
-
A peptidoglycan fragment triggers β-lactam resistance in Bacillus licheniformis.PLoS Pathog. 2012;8(3):e1002571. doi: 10.1371/journal.ppat.1002571. Epub 2012 Mar 15. PLoS Pathog. 2012. PMID: 22438804 Free PMC article.
-
Probing the Folding-Unfolding Transition of a Thermophilic Protein, MTH1880.PLoS One. 2016 Jan 14;11(1):e0145853. doi: 10.1371/journal.pone.0145853. eCollection 2016. PLoS One. 2016. PMID: 26766214 Free PMC article.
-
Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.Nucleic Acids Res. 2007;35(13):4384-95. doi: 10.1093/nar/gkm448. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576674 Free PMC article.
-
Guanidine hydrochloride-induced unfolding of the three heme coordination states of the CO-sensing transcription factor, CooA.Biochemistry. 2009 Jul 21;48(28):6585-97. doi: 10.1021/bi801827j. Biochemistry. 2009. PMID: 19594171 Free PMC article.
References
-
- Charlier P., Coyette J., Dehareng D., Dive G., Duez C., Dusart J., Fonze E., Fraipont C., Frère J. M., Galleni M., et al. Résistance bacterienne aux β-lactamases. Medecine/sciences. 1998;14:544–549.
-
- Joris B., Hardt K., Ghuysen J. M. Induction of β-lactamase and low-affinity penicillin binding protein 2′ synthesis in Gram-positive bacteria. New Compr. Biochem. 1994;27:505–515.
-
- Filée P., Benlafaya K., Delmarcelle M., Moutzourelis G., Frère J. M., Brans A., Joris B. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP β-lactamase. Mol. Microbiol. 2002;44:685–694. - PubMed
-
- Erratum. J. Bacteriol. 1987;169:3392.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

