C-type lectin-like domains in Fugu rubripes
- PMID: 15285787
- PMCID: PMC514892
- DOI: 10.1186/1471-2164-5-51
C-type lectin-like domains in Fugu rubripes
Abstract
Background: Members of the C-type lectin domain (CTLD) superfamily are metazoan proteins functionally important in glycoprotein metabolism, mechanisms of multicellular integration and immunity. Three genome-level studies on human, C. elegans and D. melanogaster reported previously demonstrated almost complete divergence among invertebrate and mammalian families of CTLD-containing proteins (CTLDcps).
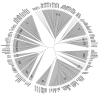
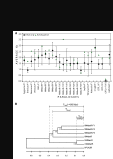
Results: We have performed an analysis of CTLD family composition in Fugu rubripes using the draft genome sequence. The results show that all but two groups of CTLDcps identified in mammals are also found in fish, and that most of the groups have the same members as in mammals. We failed to detect representatives for CTLD groups V (NK cell receptors) and VII (lithostathine), while the DC-SIGN subgroup of group II is overrepresented in Fugu. Several new CTLD-containing genes, highly conserved between Fugu and human, were discovered using the Fugu genome sequence as a reference, including a CSPG family member and an SCP-domain-containing soluble protein. A distinct group of soluble dual-CTLD proteins has been identified, which may be the first reported CTLDcp group shared by invertebrates and vertebrates. We show that CTLDcp-encoding genes are selectively duplicated in Fugu, in a manner that suggests an ancient large-scale duplication event. We have verified 32 gene structures and predicted 63 new ones, and make our annotations available through a distributed annotation system (DAS) server http://anz.anu.edu.au:8080/Fugu_rubripes/ and their sequences as additional files with this paper.
Conclusions: The vertebrate CTLDcp family was essentially formed early in vertebrate evolution and is completely different from the invertebrate families. Comparison of fish and mammalian genomes revealed three groups of CTLDcps and several new members of the known groups, which are highly conserved between fish and mammals, but were not identified in the study using only mammalian genomes. Despite limitations of the draft sequence, the Fugu rubripes genome is a powerful instrument for gene discovery and vertebrate evolutionary analysis. The composition of the CTLDcp superfamily in fish and mammals suggests that large-scale duplication events played an important role in the evolution of vertebrates.
Figures



Similar articles
-
Origin and diversity of the SOX transcription factor gene family: genome-wide analysis in Fugu rubripes.Gene. 2004 Mar 17;328:177-86. doi: 10.1016/j.gene.2003.12.008. Gene. 2004. PMID: 15019997
-
Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes.Genome Res. 2004 Jul;14(7):1248-57. doi: 10.1101/gr.2400004. Epub 2004 Jun 14. Genome Res. 2004. PMID: 15197168 Free PMC article.
-
Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts.BMC Evol Biol. 2007 Mar 30;7:49. doi: 10.1186/1471-2148-7-49. BMC Evol Biol. 2007. PMID: 17394664 Free PMC article.
-
Intermedin, a novel calcitonin family peptide that exists in teleosts as well as in mammals: a comparison with other calcitonin/intermedin family peptides in vertebrates.Peptides. 2004 Oct;25(10):1633-42. doi: 10.1016/j.peptides.2004.05.021. Peptides. 2004. PMID: 15476930 Review.
-
The C-type lectin-like domain superfamily.FEBS J. 2005 Dec;272(24):6179-217. doi: 10.1111/j.1742-4658.2005.05031.x. FEBS J. 2005. PMID: 16336259 Review.
Cited by
-
Immune-related, lectin-like receptors are differentially expressed in the myeloid and lymphoid lineages of zebrafish.Immunogenetics. 2006 Feb;58(1):31-40. doi: 10.1007/s00251-005-0064-3. Epub 2006 Feb 9. Immunogenetics. 2006. PMID: 16467987
-
Studies Into β-Glucan Recognition in Fish Suggests a Key Role for the C-Type Lectin Pathway.Front Immunol. 2019 Feb 26;10:280. doi: 10.3389/fimmu.2019.00280. eCollection 2019. Front Immunol. 2019. PMID: 30863400 Free PMC article.
-
HumanLectome, an update of UniLectin for the annotation and prediction of human lectins.Nucleic Acids Res. 2024 Jan 5;52(D1):D1683-D1693. doi: 10.1093/nar/gkad905. Nucleic Acids Res. 2024. PMID: 37889052 Free PMC article.
-
The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities.Immunogenetics. 2011 Mar;63(3):123-41. doi: 10.1007/s00251-010-0506-4. Epub 2010 Dec 30. Immunogenetics. 2011. PMID: 21191578 Free PMC article. Review.
-
Reconstructing immune phylogeny: new perspectives.Nat Rev Immunol. 2005 Nov;5(11):866-79. doi: 10.1038/nri1712. Nat Rev Immunol. 2005. PMID: 16261174 Free PMC article. Review.
References
-
- Drickamer K. Evolution of Ca(2+)-dependent animal lectins. Prog Nucleic Acid Res Mol Biol. 1993;45:207–232. - PubMed
-
- Sano H, Kuroki Y, Honma T, Ogasawara Y, Sohma H, Voelker DR, Akino T. Analysis of chimeric proteins identifies the regions in the carbohydrate recognition domains of rat lung collectins that are essential for interactions with phospholipids, glycolipids, and alveolar type II cells. J Biol Chem. 1998;273:4783–4789. doi: 10.1074/jbc.273.8.4783. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

