Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme
- PMID: 15286283
- PMCID: PMC511024
- DOI: 10.1073/pnas.0404482101
Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme
Abstract
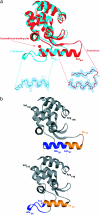
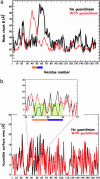
We have designed a molecular switch in a T4 lysozyme construct that controls a large-scale translation of a duplicated helix. As shown by crystal structures of the construct with the switch on and off, the conformational change is triggered by the binding of a ligand (guanidinium ion) to a site that in the wild-type protein was occupied by the guanidino head group of an Arg. In the design template, a duplicated helix is flanked by two loop regions of different stabilities. In the "on" state, the N-terminal loop is weakly structured, whereas the C-terminal loop has a well defined conformation that is stabilized by means of nonbonded interactions with the Arg head group. The truncation of the Arg to Ala destabilizes this loop and switches the protein to the "off" state, in which the duplicated helix is translocated approximately 20 A. Guanidinium binding restores the key interactions, restabilizes the C-terminal loop, and restores the "on" state. Thus, the presence of an external ligand, which is unrelated to the catalytic activity of the enzyme, triggers the inserted helix to translate 20 A away from the binding site. The results illustrate a proposed mechanism for protein evolution in which sequence duplication followed by point mutation can lead to the establishment of new function.
Figures

Similar articles
-
Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.Protein Sci. 2006 Apr;15(4):853-61. doi: 10.1110/ps.052020606. Protein Sci. 2006. PMID: 16600969 Free PMC article.
-
Molecular dynamics simulations of an engineered T4 lysozyme exclude helix to sheet transition, and provide insights into long distance, intra-protein switchable motion.Protein Sci. 2020 Feb;29(2):542-554. doi: 10.1002/pro.3780. Epub 2019 Nov 21. Protein Sci. 2020. PMID: 31702853 Free PMC article.
-
Accommodation of amino acid insertions in an alpha-helix of T4 lysozyme. Structural and thermodynamic analysis.J Mol Biol. 1994 Feb 25;236(3):869-86. doi: 10.1006/jmbi.1994.1195. J Mol Biol. 1994. PMID: 8114100
-
Size versus polarizability in protein-ligand interactions: binding of noble gases within engineered cavities in phage T4 lysozyme.J Mol Biol. 2000 Sep 29;302(4):955-77. doi: 10.1006/jmbi.2000.4063. J Mol Biol. 2000. PMID: 10993735
-
Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.J Mol Biol. 1998 Mar 27;277(2):467-85. doi: 10.1006/jmbi.1997.1606. J Mol Biol. 1998. PMID: 9514755
Cited by
-
Ligand binding and allostery can emerge simultaneously.Protein Sci. 2007 May;16(5):929-37. doi: 10.1110/ps.062706007. Epub 2007 Mar 30. Protein Sci. 2007. PMID: 17400921 Free PMC article.
-
Emerging area: biomaterials that mimic and exploit protein motion.Soft Matter. 2011 Apr;7(8):3679-3688. doi: 10.1039/C0SM01351J. Soft Matter. 2011. PMID: 25214879 Free PMC article.
-
Beyond molecular beacons: optical sensors based on the binding-induced folding of proteins and polypeptides.Chemistry. 2009;15(10):2244-51. doi: 10.1002/chem.200701748. Chemistry. 2009. PMID: 19191230 Free PMC article.
-
Protein stabilization by specific binding of guanidinium to a functional arginine-binding surface on an SH3 domain.Protein Sci. 2006 Jan;15(1):162-70. doi: 10.1110/ps.051829106. Protein Sci. 2006. PMID: 16373478 Free PMC article.
-
Designing redox potential-controlled protein switches based on mutually exclusive proteins.Protein Sci. 2012 Aug;21(8):1222-30. doi: 10.1002/pro.2109. Protein Sci. 2012. PMID: 22733630 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

