Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1
- PMID: 15286807
- PMCID: PMC484980
- DOI: 10.1172/JCI21100
Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1
Abstract
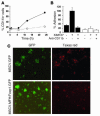
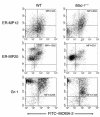
The precise signals responsible for differentiation of blood-borne monocytes into tissue macrophages are incompletely defined. "Outside-in" signaling by integrins has been implicated in modulation of gene expression that affects cellular differentiation. Herein, using differential display PCR, we have cloned an 85-kDa forkhead transcription factor (termed Mac-1-regulated forkhead [MFH] and found subsequently to be identical to Foxp1) that is downregulated in beta(2)-integrin Mac-1-clustered compared with Mac-1-nonclustered monocytic THP-1 cells. MFH/Foxp1 is expressed in untreated HL60 cells, and its expression was markedly reduced during phorbol ester-induced monocyte differentiation, but not retinoic acid-induced granulocyte differentiation. Overexpression of MFH/Foxp1 markedly attenuated phorbol ester-induced expression of c-fms, which encodes the M-CSF receptor and is obligatory for macrophage differentiation. This was accompanied by decreased CD11b expression, cell adhesiveness, and phagocytosis. Using electromobility shift and reporter assays, we have established that MFH/Foxp1 binds to previously uncharacterized sites within the c-fms promoter and functions as a transcriptional repressor. Deficiency of Mac-1 is associated with altered regulation of MFH/Foxp1 and monocyte maturation in vivo. Taken together, these observations suggest that Mac-1 engagement orchestrates monocyte-differentiation signals by regulating the expression of the forkhead transcription repressor MFH/Foxp1. This represents a new pathway for integrin-dependent modulation of gene expression and control of cellular differentiation.
Figures






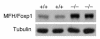

Similar articles
-
Integrin signals, transcription factors, and monocyte differentiation.Trends Cardiovasc Med. 2006 Jul;16(5):146-52. doi: 10.1016/j.tcm.2006.03.002. Trends Cardiovasc Med. 2006. PMID: 16781947 Review.
-
Leukocyte integrin signaling regulates FOXP1 gene expression via FOXP1-IT1 long non-coding RNA-mediated IRAK1 pathway.Biochim Biophys Acta Gene Regul Mech. 2019 Apr;1862(4):493-508. doi: 10.1016/j.bbagrm.2019.02.008. Epub 2019 Mar 2. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30831269 Free PMC article.
-
Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function.Blood. 2008 Dec 1;112(12):4699-711. doi: 10.1182/blood-2008-01-137018. Epub 2008 Sep 17. Blood. 2008. PMID: 18799727 Free PMC article.
-
Integrin alphaMbeta2 clustering triggers phosphorylation and activation of protein kinase C delta that regulates transcription factor Foxp1 expression in monocytes.J Immunol. 2010 Apr 1;184(7):3697-709. doi: 10.4049/jimmunol.0903316. Epub 2010 Feb 26. J Immunol. 2010. PMID: 20190138
-
Leukocyte integrin Mac-1 recruits toll/interleukin-1 receptor superfamily signaling intermediates to modulate NF-kappaB activity.Circ Res. 2001 Nov 9;89(10):859-65. doi: 10.1161/hh2201.099166. Circ Res. 2001. PMID: 11701612
Cited by
-
Cooperative regulation in development by SMRT and FOXP1.Genes Dev. 2008 Mar 15;22(6):740-5. doi: 10.1101/gad.1637108. Genes Dev. 2008. PMID: 18347093 Free PMC article.
-
A role of kindlin-3 in integrin αMβ2 outside-in signaling and the Syk-Vav1-Rac1/Cdc42 signaling axis.PLoS One. 2013;8(2):e56911. doi: 10.1371/journal.pone.0056911. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437269 Free PMC article.
-
The homeobox transcription factor VentX controls human macrophage terminal differentiation and proinflammatory activation.J Clin Invest. 2011 Jul;121(7):2599-613. doi: 10.1172/JCI45556. Epub 2011 Jun 13. J Clin Invest. 2011. PMID: 21670496 Free PMC article.
-
Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis.Genetics. 2007 Sep;177(1):631-53. doi: 10.1534/genetics.107.078584. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660535 Free PMC article.
-
NaCl potentiates human fibrocyte differentiation.PLoS One. 2012;7(9):e45674. doi: 10.1371/journal.pone.0045674. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23029177 Free PMC article.
References
-
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. - PubMed
-
- Dai XM, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. - PubMed
-
- Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J. Leukoc. Biol. 1998;63:405–417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

