Function and evolution of the serotonin-synthetic bas-1 gene and other aromatic amino acid decarboxylase genes in Caenorhabditis
- PMID: 15287963
- PMCID: PMC514703
- DOI: 10.1186/1471-2148-4-24
Function and evolution of the serotonin-synthetic bas-1 gene and other aromatic amino acid decarboxylase genes in Caenorhabditis
Abstract
Background: Aromatic L-amino acid decarboxylase (AADC) enzymes catalyze the synthesis of biogenic amines, including the neurotransmitters serotonin and dopamine, throughout the animal kingdom. These neurotransmitters typically perform important functions in both the nervous system and other tissues, as illustrated by the debilitating conditions that arise from their deficiency. Studying the regulation and evolution of AADC genes is therefore desirable to further our understanding of how nervous systems function and evolve.
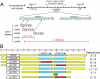
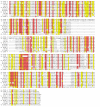
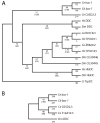
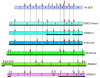
Results: In the nematode C. elegans, the bas-1 gene is required for both serotonin and dopamine synthesis, and maps genetically near two AADC-homologous sequences. We show by transformation rescue and sequencing of mutant alleles that bas-1 encodes an AADC enzyme. Expression of a reporter construct in transgenics suggests that the bas-1 gene is expressed, as expected, in identified serotonergic and dopaminergic neurons. The bas-1 gene is one of six AADC-like sequences in the C. elegans genome, including a duplicate that is immediately downstream of the bas-1 gene. Some of the six AADC genes are quite similar to known serotonin- and dopamine-synthetic AADC's from other organisms whereas others are divergent, suggesting previously unidentified functions. In comparing the AADC genes of C. elegans with those of the congeneric C. briggsae, we find only four orthologous AADC genes in C. briggsae. Two C. elegans AADC genes - those most similar to bas-1 - are missing from C. briggsae. Phylogenetic analysis indicates that one or both of these bas-1-like genes were present in the common ancestor of C. elegans and C. briggsae, and were retained in the C. elegans line, but lost in the C. briggsae line. Further analysis of the two bas-1-like genes in C. elegans suggests that they are unlikely to encode functional enzymes, and may be expressed pseudogenes.
Conclusions: The bas-1 gene of C. elegans encodes a serotonin- and dopamine-synthetic AADC enzyme. Two C. elegans AADC-homologous genes that are closely related to bas-1 are missing from the congeneric C. briggsae; one or more these genes was present in the common ancestor of C. elegans and C. briggsae. Despite their persistence in C. elegans, evidence suggests the bas-1-like genes do not encode functional AADC proteins. The presence of the genes in C. elegans raises questions about how many 'predicted genes' in sequenced genomes are functional, and how duplicate genes are retained or lost during evolution. This is another example of unexpected retention of duplicate genes in eukaryotic genomes.
Figures



Similar articles
-
The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae.J Mol Biol. 1993 Feb 20;229(4):890-908. doi: 10.1006/jmbi.1993.1094. J Mol Biol. 1993. PMID: 8445654
-
Homologs of the Caenorhabditis elegans masculinizing gene her-1 in C. briggsae and the filarial parasite Brugia malayi.Genetics. 1999 Aug;152(4):1573-84. doi: 10.1093/genetics/152.4.1573. Genetics. 1999. PMID: 10430584 Free PMC article.
-
Conservation of sequence and function of the pag-3 genes from C. elegans and C. briggsae.Gene. 2000 Feb 8;243(1-2):67-74. doi: 10.1016/s0378-1119(99)00560-0. Gene. 2000. PMID: 10675614
-
Comparative genomics in C. elegans, C. briggsae, and other Caenorhabditis species.Methods Mol Biol. 2006;351:13-29. doi: 10.1385/1-59745-151-7:13. Methods Mol Biol. 2006. PMID: 16988423 Review.
-
[Molecular genetics of aromatic L-amino acid decarboxylase].Yakubutsu Seishin Kodo. 1993 Aug;13(4):251-6. Yakubutsu Seishin Kodo. 1993. PMID: 8237142 Review. Japanese.
Cited by
-
A cocoa peptide protects Caenorhabditis elegans from oxidative stress and β-amyloid peptide toxicity.PLoS One. 2013 May 13;8(5):e63283. doi: 10.1371/journal.pone.0063283. Print 2013. PLoS One. 2013. PMID: 23675471 Free PMC article.
-
Regulation and modulation of biogenic amine neurotransmission in Drosophila and Caenorhabditis elegans.Front Physiol. 2023 Feb 16;14:970405. doi: 10.3389/fphys.2023.970405. eCollection 2023. Front Physiol. 2023. PMID: 36875033 Free PMC article. Review.
-
Dopamine signaling regulates fat content through β-oxidation in Caenorhabditis elegans.PLoS One. 2014 Jan 22;9(1):e85874. doi: 10.1371/journal.pone.0085874. eCollection 2014. PLoS One. 2014. PMID: 24465759 Free PMC article.
-
Multiple doublesex-related genes specify critical cell fates in a C. elegans male neural circuit.PLoS One. 2011;6(11):e26811. doi: 10.1371/journal.pone.0026811. Epub 2011 Nov 1. PLoS One. 2011. PMID: 22069471 Free PMC article.
-
DOPA Decarboxylase Modulates Tau Toxicity.Biol Psychiatry. 2018 Mar 1;83(5):438-446. doi: 10.1016/j.biopsych.2017.06.007. Epub 2017 Jun 15. Biol Psychiatry. 2018. PMID: 28751068 Free PMC article.
References
-
- Deakin JF. Depression and antisocial personality disorder: two contrasting disorders of 5HT function. J Neural Transm Suppl. 2003;64:79–93. - PubMed
-
- Wright TRF. The genetics of biogenic amine metabolism, sclerotization, and melanizaton in Drosophila melaogaster. Adv Genet. 1987;24:127–222. - PubMed
-
- Poupon A, Jebai F, labesse G, Gros F, Thibault J, Mornon J, Krieger M. Structure modelling and site-directed mutagenesis of the rat aromatic L-amino acid pyridoxal 5'-phosphate-dependent decarboxylase: a functional study. Proteins. 1999;37:191–203. doi: 10.1002/(SICI)1097-0134(19991101)37:2<191::AID-PROT5>3.0.CO;2-4. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

