Origins of chromosomal rearrangement hotspots in the human genome: evidence from the AZFa deletion hotspots
- PMID: 15287977
- PMCID: PMC507880
- DOI: 10.1186/gb-2004-5-8-r55
Origins of chromosomal rearrangement hotspots in the human genome: evidence from the AZFa deletion hotspots
Abstract
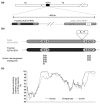
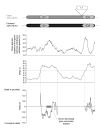
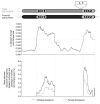
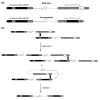
Background: The origins of the recombination hotspots that are a common feature of both allelic and non-allelic homologous recombination in the human genome are poorly understood. We have investigated, by comparative sequencing, the evolution of two hotspots of non-allelic homologous recombination on the Y chromosome that lie within paralogous sequences known to sponsor deletions resulting in male infertility.
Results: These recombination hotspots are characterized by signatures of concerted evolution, which indicate that gene conversion between paralogs has been predominant in shaping their recent evolution. By contrast, the paralogous sequences that surround the hotspots exhibit little evidence of gene conversion. A second feature of these rearrangement hotspots is the extreme interspecific sequence divergence (around 2.5%) that places them among the most divergent orthologous sequences between humans and chimpanzees.
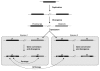
Conclusions: Several hominid-specific gene conversion events have rendered these hotspots better substrates for chromosomal rearrangements in humans than in chimpanzees or gorillas. Monte Carlo simulations of sequence evolution suggest that extreme sequence divergence is a direct consequence of gene conversion between paralogs. We propose that the coincidence of signatures of concerted evolution and recurrent breakpoints of chromosomal rearrangement (mapped at the sequence level) may enable the identification of putative rearrangement hotspots from analysis of comparative sequences from great apes.
Figures





References
-
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996;12:288–297. - PubMed
-
- Jobling MA, Williams G, Schiebel K, Pandya A, McElreavey K, Salas L, Rappold GA, Affara NA, Tyler-Smith C. A selective difference between human Y-chromosomal DNA haplotypes. Curr Biol. 1998;8:1391–1394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

