The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2
- PMID: 15289463
- PMCID: PMC514179
- DOI: 10.1101/gad.1220104
The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2
Abstract
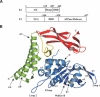
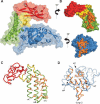
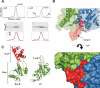
DNA replication of the papillomaviruses is specified by cooperative binding of two proteins to the ori site: the enhancer E2 and the viral initiator E1, a distant member of the AAA+ family of proteins. Formation of this prereplication complex is an essential step toward the construction of a functional, multimeric E1 helicase and DNA melting. To understand how E2 interacts with E1 to regulate this process, we have solved the X-ray structure of a complex containing the HPV18 E2 activation domain bound to the helicase domain of E1. Modeling the monomers of E1 to a hexameric helicase shows that E2 blocks hexamerization of E1 by shielding a region of the E1 oligomerization surface and stabilizing a conformation of E1 that is incompatible with ATP binding. Further biochemical experiments and structural analysis show that ATP is an allosteric effector of the dissociation of E2 from E1. Our data provide the first molecular insights into how a protein can regulate the assembly of an oligomeric AAA+ complex and explain at a structural level why E2, after playing a matchmaker role by guiding E1 to the DNA, must dissociate for subsequent steps of initiation to occur. Building on previously proposed ideas, we discuss how our data advance current models for the conversion of E1 in the prereplication complex to a hexameric helicase assembly.
Figures





References
-
- Alexandrov A.I., Botchan, M.R., and Cozzarelli, N.R. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J. Biol. Chem. 277: 44886-44897. - PubMed
-
- Antson A.A., Burns, J.E., Moroz, O.V., Scott, D.J., Sanders, C.M., Bronstein, I.B., Dodson, G.G., Wilson, K.S., and Maitland, N.J. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403: 805-809. - PubMed
-
- Bosch F.X., Manos, M.M., Munoz, N., Sherman, M., Jansen, A.M., Peto, J., Schiffman, M.H., Moreno, V., Kurman, R., and Shah, K.V. 1995. Prevalence of human papillomavirus in cervical cancer: A worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl. Cancer Inst. 87: 796-802. - PubMed
-
- Bullock P.A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32: 503-568. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
