Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia
- PMID: 15289474
- PMCID: PMC509262
- DOI: 10.1101/gr.2256604
Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia
Abstract
The Giardia lamblia genome sequencing project affords us a unique opportunity to conduct comparative analyses of core cellular systems between early and late-diverging eukaryotes on a genome-wide scale. We report a survey to identify canonical transcription components in Giardia, focusing on RNA polymerase (RNAP) subunits and transcription-initiation factors. Our survey revealed that Giardia contains homologs to 21 of the 28 polypeptides comprising eukaryal RNAPI, RNAPII, and RNAPIII; six of the seven RNAP subunits without giardial homologs are polymerase specific. Components of only four of the 12 general transcription initiation factors have giardial homologs. Surprisingly, giardial TATA-binding protein (TBP) is highly divergent with respect to archaeal and higher eukaryotic TBPs, and a giardial homolog of transcription factor IIB was not identified. We conclude that Giardia represents a transition during the evolution of eukaryal transcription systems, exhibiting a relatively complete set of RNAP subunits and a rudimentary basal initiation apparatus for each transcription system. Most class-specific RNAP subunits and basal initiation factors appear to have evolved after the divergence of Giardia from the main eukaryotic line of descent. Consequently, Giardia is predicted to be unique in many aspects of transcription initiation with respect to paradigms derived from studies in crown eukaryotes.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
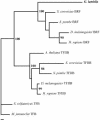
References
-
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J.D. 1994. Molecular Biology of the Cell, Third Edition. Garland Publishing, Inc. New York.
-
- Awrey, D.E., Weilbaecher, R.G., Hemming, S.A., Orlicky, S.M., Kane, C.M., and Edwards, A.M. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272: 14747–14754. - PubMed
-
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16: 512–524. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/Entrez/; Entrez Query Page, National Center for Biotechnology Information.
-
- http://rdp.cme.msu.edu/html/; Ribosomal Database Project homepage.
-
- http://hmmer.wustl.edu/; HMMER Homepage.
-
- http://www.ebi.ac.uk/clustalw; European Bioinformatics Institute ClustalW page.
-
- http://evolution.genetics.washington.edu/phylip.html; PHYLIP homepage.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
