Large-scale validation of single nucleotide polymorphisms in gene regions
- PMID: 15289484
- PMCID: PMC509276
- DOI: 10.1101/gr.2421604
Large-scale validation of single nucleotide polymorphisms in gene regions
Abstract
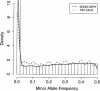
Genome-wide association studies using large numbers of bi-allelic single nucleotide polymorphisms (SNPs) have been proposed as a potentially powerful method for identifying genes involved in common diseases. To assemble a SNP collection appropriate for large-scale association, we designed assays for 226,099 publicly available SNPs located primarily within known and predicted gene regions. Allele frequencies were estimated in a sample of 92 CEPH Caucasians using chip-based MALDI-TOF mass spectrometry with pooled DNA. Of the 204,200 designed assays that were functional, 125,799 SNPs were determined to be polymorphic (minor allele frequency > 0.02), of which 101,729 map uniquely to the human genome. Many of the commonly available RefSNP annotations were predictive of polymorphic status and could be used to improve the selection of SNPs from the public domain for genetic research. The set of uniquely mapping, polymorphic SNPs is located within 10 kb of 66% of known and predicted genes annotated in LocusLink, which could prove useful for large-scale disease association studies.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures
References
-
- Altshuler, D., Pollara, V.J., Cowles, C.R., Van Etten, W.J., Baldwin, J., Linton, L., and Lander, E.S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516. - PubMed
-
- Buetow, K.H., Edmonson, M.N., and Cassidy, A.B. 1999. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21: 323–325. - PubMed
-
- Buetow, K.H., Edmonson, M., MacDonald, R., Clifford, R., Yip, P., Kelley, J., Little, D.P., Strausberg, R., Koester, H., Cantor, C.R., et al. 2001. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl. Acad. Sci. 98: 581–584. - PMC - PubMed
-
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518–521. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/SNP/; NCBI dbSNP home page.
-
- http://snp.cshl.org; The SNP Consortium home page.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
