Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape
- PMID: 15289502
- PMCID: PMC2211982
- DOI: 10.1084/jem.20040638
Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape
Abstract
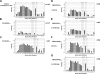
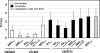
Escape mutations are believed to be important contributors to immune evasion by rapidly evolving viruses such as hepatitis C virus (HCV). We show that the majority of HCV-specific cytotoxic T lymphocyte (CTL) responses directed against viral epitopes that escaped immune recognition in HCV-infected chimpanzees displayed a reduced CDR3 amino acid diversity when compared with responses in which no CTL epitope variation was detected during chronic infection or with those associated with protective immunity. Decreased T cell receptor (TCR) CDR3 amino acid diversity in chronic infection could be detected long before the appearance of viral escape mutations in the plasma. In both chronic and resolved infection, identical T cell receptor clonotypes were present in liver and peripheral blood. These findings provide a deeper understanding of the evolution of CTL epitope variations in chronic viral infections and highlight the importance of the generation and maintenance of a diverse TCR repertoire directed against individual epitopes.
Figures




References
-
- Lauer, G.M., and B.D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52. - PubMed
-
- Cooper, S., A.L. Erickson, E.J. Adams, J. Kansopon, A.J. Weiner, D.Y. Chien, M. Houghton, P. Parham, and C.M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity. 10:439–449. - PubMed
-
- Gerlach, J.T., H.M. Diepolder, M.C. Jung, N.H. Gruener, W.W. Schraut, R. Zachoval, R. Hoffmann, C.A. Schirren, T. Santantonio, and G.R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 117:933–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

