Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V
- PMID: 15289580
- PMCID: PMC506822
- DOI: 10.1093/nar/gkh747
Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V
Abstract
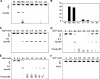
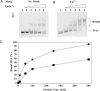
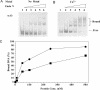
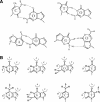
Oxanine (O) is a deamination product derived from guanine with the nitrogen at the N1 position substituted by oxygen. Cytosine, thymine, adenine, guanine as well as oxanine itself can be incorporated by Klenow Fragment to pair with oxanine in a DNA template with similar efficiency, indicating that oxanine in DNA may cause various mutations. As a nucleotide, deoxyoxanosine may substitute for deoxyguanosine to complete a primer extension reaction. Endonuclease V, an enzyme known for its enzymatic activity on uridine-, inosine- and xanthosine-containing DNA, can cleave oxanosine-containing DNA at the second phosphodiester bond 3' to the lesion. Mg2+ or Mn2+, and to a small extent Co2+ or Ni2+, support the oxanosine-containing DNA cleavage activity. All four oxanosine-containing base pairs (A/O, T/O, C/O and G/O) were cleaved with similar efficiency. The cleavage of double-stranded oxanosine-containing DNA was approximately 6-fold less efficient than that of double-stranded inosine-containing DNA. Single-stranded oxanosine-containing DNA was cleaved with a lower efficiency as compared with double-stranded oxanosine-containing DNA. A metal ion enhances the binding of endonuclease V to double-stranded and single-stranded oxanosine-containing DNA 6- and 4-fold, respectively. Hypothetic models of oxanine-containing base pairs and deaminated base recognition mechanism are presented.
Figures




Similar articles
-
Active site plasticity of endonuclease V from Salmonella typhimurium.Biochemistry. 2005 Jan 18;44(2):675-83. doi: 10.1021/bi048752j. Biochemistry. 2005. PMID: 15641793
-
Oxanine DNA glycosylase activity from Mammalian alkyladenine glycosylase.J Biol Chem. 2004 Sep 10;279(37):38177-83. doi: 10.1074/jbc.M405882200. Epub 2004 Jul 7. J Biol Chem. 2004. PMID: 15247209
-
Synthesis of 2'-deoxyoxanosine from 2'-deoxyguanosine, conversion to its phosphoramidite, and incorporation into oxanine-containing oligodeoxynucleotides.Curr Protoc Nucleic Acid Chem. 2010 Jun;Chapter 4:Unit 4.39. doi: 10.1002/0471142700.nc0439s41. Curr Protoc Nucleic Acid Chem. 2010. PMID: 20517989
-
Diversity of Endonuclease V: From DNA Repair to RNA Editing.Biomolecules. 2015 Sep 24;5(4):2194-206. doi: 10.3390/biom5042194. Biomolecules. 2015. PMID: 26404388 Free PMC article. Review.
-
Structural basis for incision at deaminated adenines in DNA and RNA by endonuclease V.Prog Biophys Mol Biol. 2015 Mar;117(2-3):134-142. doi: 10.1016/j.pbiomolbio.2015.03.005. Epub 2015 Mar 28. Prog Biophys Mol Biol. 2015. PMID: 25824682 Review.
Cited by
-
Crystal structure of E. coli endonuclease V, an essential enzyme for deamination repair.Sci Rep. 2015 Aug 5;5:12754. doi: 10.1038/srep12754. Sci Rep. 2015. PMID: 26244280 Free PMC article.
-
Switching base preferences of mismatch cleavage in endonuclease V: an improved method for scanning point mutations.Nucleic Acids Res. 2007;35(1):e2. doi: 10.1093/nar/gkl916. Epub 2006 Nov 27. Nucleic Acids Res. 2007. PMID: 17130153 Free PMC article.
-
EndoVIA for quantifying A-to-I editing and mapping the subcellular localization of edited transcripts.Methods Enzymol. 2025;710:99-130. doi: 10.1016/bs.mie.2024.11.029. Epub 2024 Dec 4. Methods Enzymol. 2025. PMID: 39870453
-
Combinatorial Domain Hunting: An effective approach for the identification of soluble protein domains adaptable to high-throughput applications.Protein Sci. 2006 Oct;15(10):2356-65. doi: 10.1110/ps.062082606. Protein Sci. 2006. PMID: 17008718 Free PMC article.
-
Chemical synthesis and thermodynamic characterization of oxanine-containing oligodeoxynucleotides.Nucleic Acids Res. 2005 Oct 11;33(18):5771-80. doi: 10.1093/nar/gki865. Print 2005. Nucleic Acids Res. 2005. PMID: 16219806 Free PMC article.
References
-
- Caulfield J.L., Wishnok,J.S. and Tannenbaum,S.R. (1998) Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem., 273, 12689–12695. - PubMed
-
- Karran P. and Lindahl,T. (1980) Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry, 19, 6005–6011. - PubMed
-
- Spencer J.P., Whiteman,M., Jenner,A. and Halliwell,B. (2000) Nitrite-induced deamination and hypochlorite-induced oxidation of DNA in intact human respiratory tract epithelial cells. Free Radic. Biol. Med., 28, 1039–1050. - PubMed
-
- Suzuki T., Yamaoka,R., Nishi,M., Ide,H. and Makino,K. (1996) Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide and calf thymus DNA treated by nitrous-acid and nitric-oxide. J. Am. Chem. Soc., 118, 2515–2516.
-
- Lucas L.T., Gatehouse,D. and Shuker,D.E. (1999) Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. A new pathway for N-nitrosocompounds to exert genotoxicity. J. Biol. Chem., 274, 18319–18326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

