Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro
- PMID: 15292140
- PMCID: PMC490933
- DOI: 10.1128/JB.186.16.5392-5399.2004
Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro
Abstract
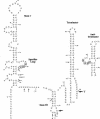
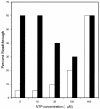
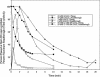
Binding of uncharged tRNA to the nascent transcript promotes readthrough of a leader region transcription termination signal in genes regulated by the T box transcription antitermination mechanism. Each gene in the T box family responds independently to its cognate tRNA, with specificity determined by base pairing of the tRNA to the leader at the anticodon and acceptor ends of the tRNA. tRNA binding stabilizes an antiterminator element in the transcript that sequesters sequences that participate in formation of the terminator helix. tRNA(Gly)-dependent antitermination of the Bacillus subtilis glyQS leader was previously demonstrated in a purified in vitro assay system. This assay system was used to investigate the kinetics of transcription through the glyQS leader and the effect of tRNA and transcription elongation factors NusA and NusG on transcriptional pausing and antitermination. Several pause sites, including a major site in the loop of stem III of the leader, were identified, and the effect of modulation of pausing on antitermination efficiency was analyzed. We found that addition of tRNA(Gly) can promote antitermination as long as the tRNA is added before the majority of the transcription complexes reach the termination site, and variations in pausing affect the requirements for timing of tRNA addition.
Figures

References
-
- Artsimovitch, I. Control of transcription termination and antitermination. In N. P. Higgins (ed.), The bacterial chromosome, in press. ASM Press, Washington, D.C.
-
- d'Aubenton Carafa, Y., E. Brody, and C. Thermes. 1990. Prediction of Rho-independent Escherichia coli transcription terminators. A statistical analysis of their stem-loop structures. J. Mol. Biol. 216:835-858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

