High salt solution structure of a left-handed RNA double helix
- PMID: 15292450
- PMCID: PMC506817
- DOI: 10.1093/nar/gkh736
High salt solution structure of a left-handed RNA double helix
Abstract
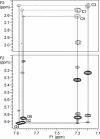
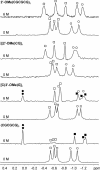
Right-handed RNA duplexes of (CG)n sequence undergo salt-induced helicity reversal, forming left-handed RNA double helices (Z-RNA). In contrast to the thoroughly studied Z-DNA, no Z-RNA structure of natural origin is known. Here we report the NMR structure of a half-turn, left-handed RNA helix (CGCGCG)2 determined in 6 M NaClO4. This is the first nucleic acid motif determined at such high salt. Sequential assignments of non-exchangeable proton resonances of the Z-form were based on the hitherto unreported NOE connectivity path [H6(n)-H5'/H5''(n)-H8(n+1)-H1'(n+1)-H6(n+2)] found for left-handed helices. Z-RNA structure shows several conformational features significantly different from Z-DNA. Intra-strand but no inter-strand base stacking was observed for both CpG and GpC steps. Helical twist angles for CpG steps have small positive values (4-7 degrees), whereas GpC steps have large negative values (-61 degrees). In the full-turn model of Z-RNA (12.4 bp per turn), base pairs are much closer to the helix axis than in Z-DNA, thus both the very deep, narrow minor groove with buried cytidine 2'-OH groups, and the major groove are well defined. The 2'-OH group of cytidines plays a crucial role in the Z-RNA structure and its formation; 2'-O-methylation of cytidine, but not of guanosine residues prohibits A to Z helicity reversal.
Figures
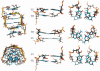

References
-
- Wang A.H.-J., Quigley,G.J., Kolpak,F.J., Crawford,J.L., van Boom,J.H., van der Marel,G. and Rich,A. (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature, 282, 680–686. - PubMed
-
- Hall K., Cruz,P., Tinoco,I.,Jr, Jovin,T.M. and van de Sande,J.H. (1984) ‘Z-RNA’. A left-handed RNA double helix. Nature, 311, 584–586. - PubMed
-
- Adamiak R.W., Galat,A. and Skalski,B. (1985) Salt- and solvent-dependent conformational transitions of ribo-CGCGCG duplex. Biochim. Biophys. Acta, 825, 345–352.
Publication types
MeSH terms
Substances
Associated data
- Actions

