Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR
- PMID: 15294789
- PMCID: PMC492386
- DOI: 10.1128/AEM.70.8.4582-4587.2004
Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR
Abstract
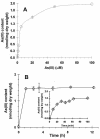
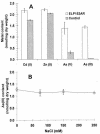
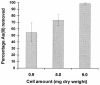
The metalloregulatory protein ArsR, which offers high affinity and selectivity toward arsenite, was overexpressed in Escherichia coli in an attempt to increase the bioaccumulation of arsenic. Overproduction of ArsR resulted in elevated levels of arsenite bioaccumulation but also a severe reduction in cell growth. Incorporation of an elastin-like polypeptide as the fusion partner to ArsR (ELP153AR) improved cell growth by twofold without compromising the ability to accumulate arsenite. Resting cells overexpressing ELP153AR accumulated 5- and 60-fold-higher levels of arsenate and arsenite than control cells without ArsR overexpression. Conversely, no significant improvement in Cd(2+) or Zn(2+) accumulation was observed, validating the specificity of ArsR. The high affinity of ArsR allowed 100% removal of 50 ppb of arsenite from contaminated water with these engineered cells, providing a technology useful to comply with the newly approved U.S. Environmental Protection Agency limit of 10 ppb. These results open up the possibility of using cells overexpressing ArsR as an inexpensive, high-affinity ligand for arsenic removal from contaminated drinking and ground water.
Figures


References
-
- Bae, W., W. Chen, A. Mulchandani, and R. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-523. - PubMed
-
- Bontidean, I., C. Berggren, G. Johansson, E. Csorgi, B. Mattiasson, J. R. Lloyd, K. J. Jakeman, and N. L. Brown. 1998. Detection of heavy metal ions at femtomolar levels using protein-based biosensors. Anal. Chem. 70:4162-4169. - PubMed
-
- Canovas, D., C. Duran, N. Rodriguez, R. Amils, and V. De Lorenzo. 2003. Testing the limits of biological tolerance to arsenic in a fungus isolated from the River Tinto. Environ. Microbiol. 5:133-138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

