Detection and quantification of the red tide dinoflagellate Karenia brevis by real-time nucleic acid sequence-based amplification
- PMID: 15294808
- PMCID: PMC492458
- DOI: 10.1128/AEM.70.8.4727-4732.2004
Detection and quantification of the red tide dinoflagellate Karenia brevis by real-time nucleic acid sequence-based amplification
Abstract
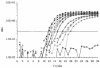
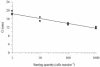
Nucleic acid sequence-based amplification (NASBA) is an isothermal method of RNA amplification that has been previously used in clinical diagnostic testing. A real-time NASBA assay has been developed for the detection of rbcL mRNA from the red tide dinoflagellate Karenia brevis. This assay is sensitive to one K. brevis cell and 1.0 fg of in vitro transcript, with occasional detection of lower concentrations of transcript. The assay did not detect rbcL mRNA from a wide range of nontarget organisms and environmental clones, while 10 strains (all tested) of K. brevis were detected. By the use of standard curves based on time to positivity, concentrations of K. brevis in environmental samples were predicted by NASBA and classified into different levels of blooms per the Florida Fish and Wildlife Conservation Commission (FWC) system. NASBA classification matched FWC classification (based on cell counts) 72% of the time. Those samples that did not match were off by only one class. NASBA is sensitive, rapid, and effective and may be used as an additional or alternative method to detect and quantify K. brevis in the marine environment.
Figures
Similar articles
-
Detection and quantification of the toxic microalgae Karenia brevis using lab on a chip mRNA sequence-based amplification.J Microbiol Methods. 2017 Aug;139:189-195. doi: 10.1016/j.mimet.2017.06.008. Epub 2017 Jun 8. J Microbiol Methods. 2017. PMID: 28602754
-
Molecular detection and quantitation of the red tide dinoflagellate Karenia brevis in the marine environment.Appl Environ Microbiol. 2003 Sep;69(9):5726-30. doi: 10.1128/AEM.69.9.5726-5730.2003. Appl Environ Microbiol. 2003. PMID: 12957971 Free PMC article.
-
Increased precision of microbial RNA quantification using NASBA with an internal control.J Microbiol Methods. 2005 Mar;60(3):343-52. doi: 10.1016/j.mimet.2004.10.011. J Microbiol Methods. 2005. PMID: 15649536
-
Characteristics and applications of nucleic acid sequence-based amplification (NASBA).Mol Biotechnol. 2002 Feb;20(2):163-79. doi: 10.1385/MB:20:2:163. Mol Biotechnol. 2002. PMID: 11876473 Review.
-
The use of NASBA for the detection of microbial pathogens in food and environmental samples.J Microbiol Methods. 2003 May;53(2):165-74. doi: 10.1016/s0167-7012(03)00022-8. J Microbiol Methods. 2003. PMID: 12654488 Review.
Cited by
-
Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR.Appl Environ Microbiol. 2006 Apr;72(4):2496-506. doi: 10.1128/AEM.72.4.2496-2506.2006. Appl Environ Microbiol. 2006. PMID: 16597949 Free PMC article.
-
Electroporation and lysis of marine microalga Karenia brevis for RNA extraction and amplification.J R Soc Interface. 2011 Apr 6;8(57):601-8. doi: 10.1098/rsif.2010.0445. Epub 2010 Nov 17. J R Soc Interface. 2011. PMID: 21084344 Free PMC article.
-
The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species.Environ Sci Pollut Res Int. 2013 Oct;20(10):6851-62. doi: 10.1007/s11356-012-1377-z. Epub 2012 Dec 18. Environ Sci Pollut Res Int. 2013. PMID: 23247526 Free PMC article.
-
Phytoplankton-group specific quantitative polymerase chain reaction assays for RuBisCO mRNA transcripts in seawater.Mar Biotechnol (NY). 2007 Nov-Dec;9(6):747-59. doi: 10.1007/s10126-007-9027-z. Epub 2007 Aug 13. Mar Biotechnol (NY). 2007. PMID: 17694413
-
Centers for Oceans and Human Health: a unified approach to the challenge of harmful algal blooms.Environ Health. 2008 Nov 7;7 Suppl 2(Suppl 2):S2. doi: 10.1186/1476-069X-7-S2-S2. Environ Health. 2008. PMID: 19025673 Free PMC article. Review.
References
-
- Adachi, M., Y. Sako, and Y. Ishida. 1996. Identification of the toxic dinoflagellates Alexandrium catenella and A. tamarense (Dinophyceae) using DNA probes and whole-cell hybridization. J. Phycol. 32:1049-1052.
-
- Culverhouse, P. F., R. Williams, B. Reguera, V. Henry, and S. Gonzalez-Gil. 2003. Do experts make mistakes? A comparison of human and machine identification of dinoflagellates. Mar. Ecol. Prog. Ser. 247:17-25.
-
- Davey, C., and L. T. Malek. 1989. Nucleic acid amplification process. European patent EP 0329822.
-
- Godhe, A., S. K. Otta, A.-S. Rehnstam-Holm, I. Karunasagar, and I. Karunasagar. 2001. Polymerase chain reaction in detection of Gymnodinium mikimotoi and Alexandrium minutum in field samples from southwest India. Mar. Biotechnol. 3:152-162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

