New strategies for cultivation and detection of previously uncultured microbes
- PMID: 15294811
- PMCID: PMC492380
- DOI: 10.1128/AEM.70.8.4748-4755.2004
New strategies for cultivation and detection of previously uncultured microbes
Abstract
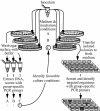
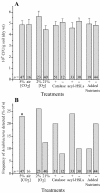
An integrative approach was used to obtain pure cultures of previously uncultivated members of the divisions Acidobacteria and Verrucomicrobia from agricultural soil and from the guts of wood-feeding termites. Some elements of the cultivation procedure included the following: the use of agar media with little or no added nutrients; relatively long periods of incubation (more than 30 days); protection of cells from exogenous peroxides; and inclusion of humic acids or a humic acid analogue (anthraquinone disulfonate) and quorum-signaling compounds (acyl homoserine lactones) in growth media. The bacteria were incubated in the presence of air and in hypoxic (1 to 2% O(2) [vol/vol]) and anoxic atmospheres. Some bacteria were incubated with elevated concentrations of CO(2) (5% [vol/vol]). Significantly more Acidobacteria were found on isolation plates that had been incubated with 5% CO(2). A simple, high-throughput, PCR-based surveillance method (plate wash PCR) was developed. This method greatly facilitated detection and ultimate isolation of target bacteria from as many as 1,000 colonies of nontarget microbes growing on the same agar plates. Results illustrate the power of integrating culture methods with molecular techniques to isolate bacteria from phylogenetic groups underrepresented in culture.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bloem, J. 1995. Fluorescent staining of microbes for total direct counts, section 4.1.8, p. 1-12. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

