Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis
- PMID: 15294817
- PMCID: PMC492348
- DOI: 10.1128/AEM.70.8.4800-4806.2004
Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis
Abstract
Denaturing gradient gel electrophoresis (DGGE) has become a widely used tool to examine microbial diversity and community structure, but no systematic comparison has been made of the DGGE profiles obtained when different hypervariable (V) regions are amplified from the same community DNA samples. We report here a study to make such comparisons and establish a preferred choice of V region(s) to examine by DGGE, when community DNA extracted from samples of digesta is used. When the members of the phylogenetically representative set of 218 rrs genes archived in the RDP II database were compared, the V1 region was found to be the most variable, followed by the V9 and V3 regions. The temperature of the lowest-melting-temperature (T(m(L))) domain for each V region was also calculated for these rrs genes, and the V1 to V4 region was found to be most heterogeneous with respect to T(m(L)). The average T(m(L)) values and their standard deviations for each V region were then used to devise the denaturing gradients suitable for separating 95% of all the sequences, and the PCR-DGGE profiles produced from the same community DNA samples with these conditions were compared. The resulting DGGE profiles were substantially different in terms of the number, resolution, and relative intensity of the amplification products. The DGGE profiles of the V3 region were best, and the V3 to V5 and V6 to V8 regions produced better DGGE profiles than did other multiple V-region amplicons. Introduction of degenerate bases in the primers used to amplify the V1 or V3 region alone did not improve DGGE banding profiles. Our results show that DGGE analysis of gastrointestinal microbiomes is best accomplished by the amplification of either the V3 or V1 region of rrs genes, but if a longer amplification product is desired, then the V3 to V5 or V6 to V8 region should be targeted.
Figures


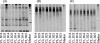
References
-
- Casamayor, E. O., H. Schafer, L. Baneras, C. Pedros-Alio, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. - PMC - PubMed
-
- Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-802. - PubMed
-
- Curtis, T. P. 1998. The comparison of the diversity of activated sludge plants. Water Sci. Technol. 37:71-78.
-
- De Boever, P., R. Wouters, V. Vermeirssen, N. Boon, and W. Verstraete. 2001. Development of a six-stage culture system for simulating the gastrointestinal microbiota of weaned infants. Microbiol. Ecol. Health Dis. 13:111-123.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

