Correspondence between community structure and function during succession in phenol- and phenol-plus-trichloroethene-fed sequencing batch reactors
- PMID: 15294835
- PMCID: PMC492464
- DOI: 10.1128/AEM.70.8.4950-4960.2004
Correspondence between community structure and function during succession in phenol- and phenol-plus-trichloroethene-fed sequencing batch reactors
Abstract
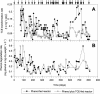
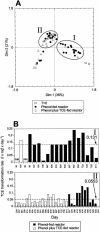
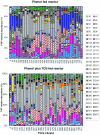
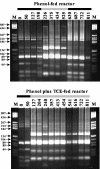
The effects of more than 2 years of trichloroethene (TCE) application on community succession and function were studied in two aerobic sequencing batch reactors. One reactor was fed phenol, and the second reactor was fed both phenol and TCE in sequence twice per day. After initiation of TCE loading in the second reactor, the TCE transformation rates initially decreased, but they stabilized with an average second-order rate coefficient of 0.044 liter mg(-1) day(-1) for 2 years. In contrast, the phenol-fed reactor showed higher and unstable TCE transformation rates, with an average rate coefficient of 0.093 liter mg(-1) day(-1). Community analysis by terminal restriction fragment length polymorphism (T-RFLP) analysis of the 16S rRNA genes showed that the phenol-plus-TCE-fed reactor had marked changes in community structure during the first 100 days and remained relatively stable afterwards, corresponding to the period of stable function. In contrast, the community structure of the phenol-fed reactor changed periodically, and the changes coincided with the periodicity observed in the TCE transformation rates. Correspondence analysis of each reactor community showed that different community structures corresponded with function (TCE degradation rate). Furthermore, the phenol hydroxylase genotypes, as determined by restriction fragment length polymorphism analysis, corresponded to community structure patterns identified by T-RFLP analysis and to periods when the TCE transformation rates were high. Long-term TCE stress appeared to select for a different and stable community structure, with lower but stable TCE degradation rates. In contrast, the community under no stress exhibited a dynamic structure and dynamic function.
Figures



References
-
- Agency for Toxic Substances and Disease Registry. 1994. National exposure registry. Trichloroethylene (TCE) subregistry baseline technical report (revised). NTIS publication PB95-154589. Agency for Toxic Substances and Disease Registry, U. S. Public Health Service, Department of Health and Human Services, Atlanta, Ga.
-
- Alexander, M. 1994. Biodegradation and bioremediation. Academic Press, San Diego, Calif.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

