GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes
- PMID: 15297299
- PMCID: PMC3037731
- DOI: 10.1093/bioinformatics/bth456
GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes
Abstract
Summary: GO::TermFinder comprises a set of object-oriented Perl modules for accessing Gene Ontology (GO) information and evaluating and visualizing the collective annotation of a list of genes to GO terms. It can be used to draw conclusions from microarray and other biological data, calculating the statistical significance of each annotation. GO::TermFinder can be used on any system on which Perl can be run, either as a command line application, in single or batch mode, or as a web-based CGI script.
Availability: The full source code and documentation for GO::TermFinder are freely available from http://search.cpan.org/dist/GO-TermFinder/.
Figures
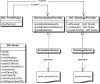

References
-
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300.
-
- Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. - PubMed
-
- Castillo-Davis CI, Hartl DL. GeneMerge–post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

