Genetic variability of the G glycoprotein gene of human metapneumovirus
- PMID: 15297494
- PMCID: PMC497612
- DOI: 10.1128/JCM.42.8.3532-3537.2004
Genetic variability of the G glycoprotein gene of human metapneumovirus
Abstract
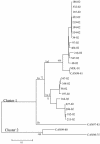
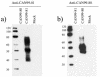
Human metapneumovirus (hMPV) has been associated with respiratory illnesses like those caused by human respiratory syncytial virus (HRSV) infection. Similar to other pneumoviruses, genetic diversity has been reported for hMPV. Little information is currently available on the genetic variability of the G glycoprotein (G), which is the most variable gene in RSV and avian pneumovirus. The complete nucleotide sequences of the G open reading frame (ORF) of 24 Canadian hMPV isolates were determined. Phylogenetic analysis showed the existence of two major groups or clusters (1 and 2). All but one of the hMPV isolates that we examined belonged to cluster 1. Additional genetic variability was observed in cluster 1, which separated into two genetic subclusters. Within cluster 1 the nucleotide sequence identity for the G ORF was 74.2 to 100%, and the identity for the predicted amino acid sequence was 61.4 to 100%. The G genes of cluster 1 isolates were more divergent from the cluster 2 isolates, with 45.6 to 50.5% and 34.2 to 37.2% identity levels for the nucleotide and amino acid sequences, respectively. Sequence analysis also revealed changes in stop codon usage, resulting in G proteins of different lengths (217, 219, 228, and 236 residues). Western blot analysis with the use of hMPV-specific polyclonal antisera to each hMPV cluster showed significant antigenic divergence between the G proteins of clusters 1 and 2. These results suggest that the G protein of hMPV is continuously evolving and that the genetic diversity observed for the hMPV genes is reflected in the antigenic variability, similar to HRSV.
Figures



References
-
- Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. - PubMed
-
- Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

