3 dimensional modelling of early human brain development using optical projection tomography
- PMID: 15298700
- PMCID: PMC514604
- DOI: 10.1186/1471-2202-5-27
3 dimensional modelling of early human brain development using optical projection tomography
Abstract
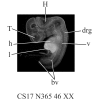
Background: As development proceeds the human embryo attains an ever more complex three dimensional (3D) structure. Analyzing the gene expression patterns that underlie these changes and interpreting their significance depends on identifying the anatomical structures to which they map and following these patterns in developing 3D structures over time. The difficulty of this task greatly increases as more gene expression patterns are added, particularly in organs with complex 3D structures such as the brain. Optical Projection Tomography (OPT) is a new technology which has been developed for rapidly generating digital 3D models of intact specimens. We have assessed the resolution of unstained neuronal structures within a Carnegie Stage (CS)17 OPT model and tested its use as a framework onto which anatomical structures can be defined and gene expression data mapped.
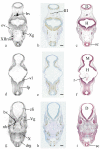
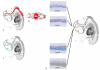
Results: Resolution of the OPT models was assessed by comparison of digital sections with physical sections stained, either with haematoxylin and eosin (H&E) or by immunocytochemistry for GAP43 or PAX6, to identify specific anatomical features. Despite the 3D models being of unstained tissue, peripheral nervous system structures from the trigeminal ganglion (approximately 300 microm by approximately 150 microm) to the rootlets of cranial nerve XII (approximately 20 microm in diameter) were clearly identifiable, as were structures in the developing neural tube such as the zona limitans intrathalamica (core is approximately 30 microm thick). Fourteen anatomical domains have been identified and visualised within the CS17 model. Two 3D gene expression domains, known to be defined by Pax6 expression in the mouse, were clearly visible when PAX6 data from 2D sections were mapped to the CS17 model. The feasibility of applying the OPT technology to all stages from CS12 to CS23, which encompasses the major period of organogenesis for the human developing central nervous system, was successfully demonstrated.
Conclusion: In the CS17 model considerable detail is visible within the developing nervous system at a minimum resolution of approximately 20 microm and 3D anatomical and gene expression domains can be defined and visualised successfully. The OPT models and accompanying technologies for manipulating them provide a powerful approach to visualising and analysing gene expression and morphology during early human brain development.
Figures
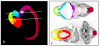

References
-
- O'Rahilly R, Muller F. The embryonic human brain An atlas of developmental stages. 2. Wiley-Liss; 1999.
-
- Desmond ME, O'Rahilly R. The growth of the human brain during the embryonic period proper. 1. Linear axes. Anat Embryol (Berl) 1981;162:137–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

