Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein
- PMID: 15299117
- PMCID: PMC520808
- DOI: 10.1104/pp.104.046086
Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein
Abstract
The P19 protein of tomato bushy stunt virus (TBSV) is a multifunctional pathogenicity determinant involved in suppression of posttranscriptional gene silencing, virus movement, and symptom induction. Here, we report that P19 interacts with the conserved RNA-binding domain of an as yet uncharacterized family of plant ALY proteins that, in animals, are involved in export of RNAs from the nucleus and transcriptional coactivation. We show that the four ALY proteins encoded by the Arabidopsis genome and two ALY proteins from Nicotiana benthamiana are localized to the nucleus. Moreover, and in contrast to animal ALY, all but one of the proteins are also in the nucleolus, with distinct subnuclear localizations. Infection of plants by TBSV or expression of P19 from Agrobacterium results in relocation of three of the six ALY proteins from the nucleus to the cytoplasm demonstrating specific targeting of the ALY proteins by P19. The differential effects on subcellular localization indicate that, in plants, the various ALY proteins may have different functions. Interaction with and relocalization of ALY is prevented by mutation of P19 at residues previously shown to be important for P19 function in plants. Down-regulation of expression of two N. benthamiana ALY genes by virus-induced gene silencing did not interfere with posttranscriptional gene silencing. Targeting of ALY proteins during TBSV infection may therefore be related to functions of P19 in addition to its silencing suppression activity.
Figures



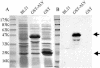

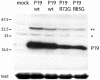
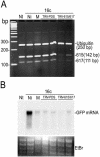
Similar articles
-
Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity.J Virol. 2006 Sep;80(18):9064-72. doi: 10.1128/JVI.00953-06. J Virol. 2006. PMID: 16940518 Free PMC article.
-
Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing.Mol Plant Microbe Interact. 2002 Mar;15(3):193-202. doi: 10.1094/MPMI.2002.15.3.193. Mol Plant Microbe Interact. 2002. PMID: 11952121
-
The multifunctional plant viral suppressor of gene silencing P19 interacts with itself and an RNA binding host protein.Virology. 2004 May 20;323(1):49-58. doi: 10.1016/j.virol.2004.02.008. Virology. 2004. PMID: 15165818
-
Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity.Mol Plant Microbe Interact. 2002 Mar;15(3):269-80. doi: 10.1094/MPMI.2002.15.3.269. Mol Plant Microbe Interact. 2002. PMID: 11952130
-
Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion.Virology. 2000 Jan 5;266(1):79-87. doi: 10.1006/viro.1999.0071. Virology. 2000. PMID: 10612662
Cited by
-
Engineering Resistance Against Viruses in Field Crops Using CRISPR- Cas9.Curr Genomics. 2021 Oct 18;22(3):214-231. doi: 10.2174/1389202922666210412102214. Curr Genomics. 2021. PMID: 34975291 Free PMC article. Review.
-
Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing.Nucleic Acids Res. 2019 Feb 28;47(4):1880-1895. doi: 10.1093/nar/gky1261. Nucleic Acids Res. 2019. PMID: 30576513 Free PMC article.
-
The cucumber mosaic virus 1a protein regulates interactions between the 2b protein and ARGONAUTE 1 while maintaining the silencing suppressor activity of the 2b protein.PLoS Pathog. 2020 Dec 3;16(12):e1009125. doi: 10.1371/journal.ppat.1009125. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33270799 Free PMC article.
-
Nuclear-cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA-silencing suppressor.J Virol. 2014 May;88(10):5228-41. doi: 10.1128/JVI.00284-14. Epub 2014 Mar 5. J Virol. 2014. PMID: 24599997 Free PMC article.
-
Nuclear export of plant pararetrovirus mRNAs involves the TREX complex, two viral proteins and the highly structured 5' leader region.Nucleic Acids Res. 2021 Sep 7;49(15):8900-8922. doi: 10.1093/nar/gkab653. Nucleic Acids Res. 2021. PMID: 34370034 Free PMC article.
References
-
- Anandalakshmi R, Marathe R, Ge X, Herr JM Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290: 142–144 - PubMed
-
- Bruhn L, Munnerlyn A, Grosschedl R (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev 11: 640–653 - PubMed
-
- Canto T, Cillo F, Palukaitis P (2002) Generation of siRNAs by T-DNA sequences does not require active transcription or homology to sequences in the plant. Mol Plant Microbe Interact 15: 1137–1146 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

