A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates
- PMID: 15299127
- PMCID: PMC520793
- DOI: 10.1104/pp.104.043398
A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates
Abstract
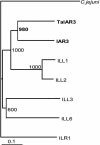
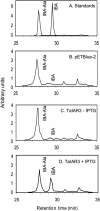
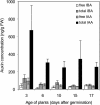
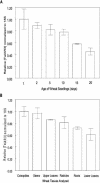
This study investigates how the ILR1-like indole acetic acid (IAA) amidohydrolase family of genes has functionally evolved in the monocotyledonous species wheat (Triticum aestivum). An ortholog for the Arabidopsis IAR3 auxin amidohydrolase gene has been isolated from wheat (TaIAR3). The TaIAR3 protein hydrolyzes negligible levels of IAA-Ala and no other IAA amino acid conjugates tested, unlike its ortholog IAR3. Instead, TaIAR3 has low specificity for the ester conjugates IAA-Glc and IAA-myoinositol and high specificity for the conjugates of indole-3-butyric acid (IBA-Ala and IBA-Gly) and indole-3-propionic-acid (IPA-Ala) so far tested. TaIAR3 did not convert the methyl esters of the IBA conjugates with Ala and Gly. IBA and IBA conjugates were detected in wheat seedlings by gas chromatography-mass spectrometry, where the conjugate of IBA with Ala may serve as a natural substrate for this enzyme. Endogenous IPA and IPA conjugates were not detected in the seedlings. Additionally, crude protein extracts of wheat seedlings possess auxin amidohydrolase activity. Temporal expression studies of TaIAR3 indicate that the transcript is initially expressed at day 1 after germination. Expression decreases through days 2, 5, 10, 15, and 20. Spatial expression studies found similar levels of expression throughout all wheat tissues examined.
Figures
References
-
- Albert VA, Oppenheimer DG, Lindqvist C (2002) Pleiotropy, redundancy and the evolution of flowers. Trends Plant Sci 7: 297–301 - PubMed
-
- Andersson B, Sandberg G (1982) Identification of endogenous N-(3-indoleacetyl)aspartic acid in Scots pine (Pinus sylvestris L.) by combined gas chromatography-mass spectrometry, using high-performance liquid chromatography for quantification. J Chromatogr 238: 151–156
-
- Applied Biosystems Incorporated (2001) User Bulletin #2: ABI PRISM 7700 Sequence Detection System. http://www.appliedbiosystems.com
-
- Aung LH (1972) The nature of root-promoting substances in Lycopersicon esculentum seedlings. Physiol Plant 26: 306–309
-
- Bandurski RS, Cohen JD, Slovin JP, Reinecke DM (1995) Auxin biosynthesis and metabolism. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 39–65
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

