AtPng1p. The first plant transglutaminase
- PMID: 15299133
- PMCID: PMC520776
- DOI: 10.1104/pp.104.042549
AtPng1p. The first plant transglutaminase
Abstract
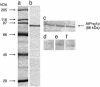
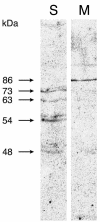
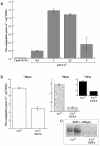
Studies have revealed in plant chloroplasts, mitochondria, cell walls, and cytoplasm the existence of transglutaminase (TGase) activities, similar to those known in animals and prokaryotes having mainly structural roles, but no protein has been associated to this type of activity in plants. A recent computational analysis has shown in Arabidopsis the presence of a gene, AtPng1p, which encodes a putative N-glycanase. AtPng1p contains the Cys-His-Asp triad present in the TGase catalytic domain. AtPng1p is a single gene expressed ubiquitously in the plant but at low levels in all light-assayed conditions. The recombinant AtPng1p protein could be immuno-detected using animal TGase antibodies. Furthermore, western-blot analysis using antibodies raised against the recombinant AtPng1p protein have lead to its detection in microsomal fraction. The purified protein links polyamines-spermine (Spm) > spermidine (Spd) > putrescine (Put)-and biotin-cadaverine to dimethylcasein in a calcium-dependent manner. Analyses of the gamma-glutamyl-derivatives revealed that the formation of covalent linkages between proteins and polyamines occurs via the transamidation of gamma-glutamyl residues of the substrate, confirming that the AtPng1p gene product acts as a TGase. The Ca(2+)- and GTP-dependent cross-linking activity of the AtPng1p protein can be visualized by the polymerization of bovine serum albumine, obtained, like the commercial TGase, at basic pH and in the presence of dithiotreitol. To our knowledge, this is the first reported plant protein, characterized at molecular level, showing TGase activity, as all its parameters analyzed so far agree with those typically exhibited by the animal TGases.
Figures


Similar articles
-
The Arabidopsis AtPNG1 gene encodes a peptide: N-glycanase.Plant J. 2007 Oct;52(1):94-104. doi: 10.1111/j.1365-313X.2007.03215.x. Epub 2007 Jul 30. Plant J. 2007. PMID: 17666024
-
Identification of mammalian-type transglutaminase in Physarum polycephalum. Evidence from the cDNA sequence and involvement of GTP in the regulation of transamidating activity.Eur J Biochem. 2002 Jul;269(14):3451-60. doi: 10.1046/j.1432-1033.2002.03026.x. Eur J Biochem. 2002. PMID: 12135484
-
Molecular cloning and characterization of a maize transglutaminase complementary DNA.Gene. 2004 Jul 7;336(1):93-104. doi: 10.1016/j.gene.2004.03.025. Gene. 2004. PMID: 15225879
-
Plant and animal transglutaminases: do similar functions imply similar structures?Amino Acids. 2009 Apr;36(4):643-57. doi: 10.1007/s00726-008-0131-9. Epub 2008 Jul 12. Amino Acids. 2009. PMID: 18622667 Review.
-
The emerging structural understanding of transglutaminase 3.J Struct Biol. 2004 Aug;147(2):200-7. doi: 10.1016/j.jsb.2004.03.009. J Struct Biol. 2004. PMID: 15193648 Review.
Cited by
-
Transglutaminases and their substrates in kinetin-stimulated etioplast-to-chloroplast transformation in cucumber cotyledons.Protoplasma. 2008 Nov;233(3-4):187-94. doi: 10.1007/s00709-008-0002-y. Epub 2008 Jun 19. Protoplasma. 2008. PMID: 18563516
-
Expression of soluble recombinant transglutaminase from Zea mays in Pichia pastoris.World J Microbiol Biotechnol. 2013 May;29(5):939-47. doi: 10.1007/s11274-012-1250-8. Epub 2013 Jan 8. World J Microbiol Biotechnol. 2013. PMID: 23296916
-
The acropetal wave of developmental cell death of tobacco corolla is preceded by activation of transglutaminase in different cell compartments.Plant Physiol. 2007 Jun;144(2):1211-22. doi: 10.1104/pp.106.092072. Epub 2007 Apr 13. Plant Physiol. 2007. PMID: 17434993 Free PMC article.
-
Fungal transglutaminase domain-containing proteins are involved in hyphal protection at the septal pore against wounding.Mol Biol Cell. 2023 Dec 1;34(13):ar127. doi: 10.1091/mbc.E23-01-0021. Epub 2023 Sep 27. Mol Biol Cell. 2023. PMID: 37756125 Free PMC article.
-
Polyamine catabolism adds fuel to leaf senescence.Amino Acids. 2017 Jan;49(1):49-56. doi: 10.1007/s00726-016-2377-y. Epub 2016 Dec 30. Amino Acids. 2017. PMID: 28039518 Free PMC article. Review.
References
-
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF (1998) Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes. Detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers. J Biol Chem 273: 3452–3460 - PubMed
-
- Bernet E, Claparols I, Dondini L, Santos MA, Serafini-Fracassini D, Torné JM (1999) Changes in polyamine content, arginine and ornithine decarboxilases and transglutaminase activities during light/dark phases of initial differentiation in maize calluses and their chloroplasts. Plant Physiol Biochem 37: 1–11
-
- Del Duca S, Bregoli AM, Bergamini C, Serafini-Fracassini D (1997) Transglutaminase-catalyzed modification of cytoskeletal proteins by polyamines during the germination of Malus domestica pollen. Sex Plant Reprod 10: 89–95
-
- Del Duca S, Dondini L, Della Mea M, Muñoz de Rueda P, Serafini-Fracassini D (2000) Factors affecting transglutaminase activity catalysing polyamine conjugation to endogenous substrates in the entire chloroplast. Plant Physiol Biochem 38: 429–439
-
- Del Duca S, Tidu V, Bassi R, Serafini-Fracassini D, Esposito C (1994) Identification of transglutaminase activity and its substrates in isolated chloroplast of Helianthus tuberosus. Planta 193: 283–289
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

