Induction and characterization of taxol-resistance phenotypes with a transiently expressed artificial transcriptional activator library
- PMID: 15304545
- PMCID: PMC514398
- DOI: 10.1093/nar/gnh114
Induction and characterization of taxol-resistance phenotypes with a transiently expressed artificial transcriptional activator library
Abstract
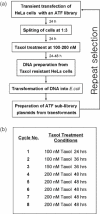
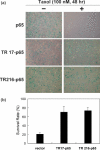
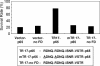
Phenotype-based functional genomic methods are useful for the identification of genes that are related to a particular biological function or disease. Essential to this approach is the ability to regulate the expression of selected genes. Artificial transcription factors (ATFs) are key molecular tools that selectively regulate gene expression in vivo. Here, we use an ATF library to identify genes that participate in rendering a cell resistant to the drug Taxol, a potent anti-cancer drug that binds to tubulin and inhibits cell division. The library, which encodes ATFs that activate (rather than inhibit) transcription, was introduced into a HeLa cell line, and Taxol-resistant cells were selected. After eight rounds of selection, we identified two ATFs that significantly increased the level of Taxol resistance (TR) in HeLa cells. Gene expression microarray experiments using these ATFs identified 37 co-regulated genes, including genes already known to participate in TR. This study demonstrates that ATF libraries can be used to induce phenotypic alterations in eukaryotic cells and then identify specific genes that are associated with the phenotype of choice.
Figures
References
-
- Berns K., Hijmans,E.M., Mullenders,J., Brummelkamp,T.R., Velds,A., Heimerikx,M., Kerkhoven,R.M., Madiredjo,M., Nijkamp,W., Weigelt,B. et al. (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature, 428, 431–437. - PubMed
-
- Suyama E., Kawasaki,H. and Taira,K. (2003) Use of a ribozyme library for validation of gene functions and cellular pathways. Nucleic Acids Res. (Suppl.), 3, 253–254. - PubMed
-
- Yin D., Fox,B., Lonetto,M.L., Etherton,M.R., Payne,D.J., Holmes,D.J., Rosenberg,M. and Ji,Y. (2004) Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics, 5, 101–113. - PubMed
-
- Kuruvilla F.G., Shamji,A.F., Sternson,S.M., Hergenrother,P.J.and Schreiber,S.L. (2002) Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature, 416, 653–657. - PubMed
-
- Michiels F., van Es,H., van Rompaey,L., Merchiers,P., Francken,B., Pittois,K., van der Schueren,J., Brys,R., Vandersmissen,J., Beirinckx,F. et al. (2002) Arrayed adenoviral expression libraries for functional screening. Nat. Biotechnol., 20, 1154–1157. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

