Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing
- PMID: 15304642
- PMCID: PMC514475
- DOI: 10.1073/pnas.0404728101
Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing
Abstract
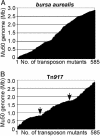
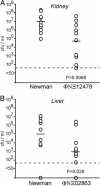
Staphylococcus aureus is the leading cause of wound and hospital-acquired infections worldwide. The emergence of S. aureus strains with resistance to multiple antibiotics requires the identification of bacterial virulence genes and the development of novel therapeutic strategies. Herein, bursa aurealis, a mariner-based transposon, was used for random mutagenesis and for the isolation of 10,325 S. aureus variants with defined insertion sites. By screening for loss-of-function mutants in a Caenorhabditis elegans killing assay, 71 S. aureus virulence genes were identified. Some of these genes are also required for S. aureus abscess formation in a murine infection model.
Figures


References
-
- Lowy, F. D. (1998) New Engl. J. Med. 339, 520–532. - PubMed
-
- Archer, G. L. (1998) Clin. Infect. Dis. 26, 1179–1181. - PubMed
-
- Gillet, Y., Issartel, B., Vanhems, P., Fournet, J. C., Lina, G., Bes, M., Vandenesch, F., Piemont, Y., Brousse, N., Floret, D. & Etienne, J. (2002) Lancet 359, 753–759. - PubMed
-
- Archer, G. L. & Climo, M. W. (2001) N. Engl. J. Med. 344, 55–56. - PubMed
-
- Neu, H. C. (1992) Science 257, 1064–1073. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

