Understanding the alpha-helix to coil transition in polypeptides using network rigidity: predicting heat and cold denaturation in mixed solvent conditions
- PMID: 15307195
- PMCID: PMC4667961
- DOI: 10.1002/bip.20102
Understanding the alpha-helix to coil transition in polypeptides using network rigidity: predicting heat and cold denaturation in mixed solvent conditions
Abstract
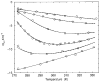
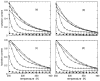
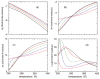
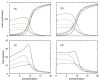
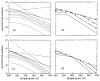
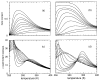
Thermodynamic stability in polypeptides is described using a novel Distance Constraint Model (DCM). Here, microscopic interactions are represented as constraints. A topological arrangement of constraints define a mechanical framework. Each constraint in the framework is associated with an enthalpic and entropic contribution. All accessible topological arrangements of distance constraints form an ensemble of mechanical frameworks, each representing a microstate of the polypeptide. A partition function is calculated exactly using a transfer matrix approach, where in many respects the DCM is similar to the Lifson-Roig model. The crucial difference is that the effect of network rigidity is explicitly calculated for each mechanical framework in the ensemble. Network rigidity is a mechanical interaction that provides a mechanism for long-range molecular cooperativity and enables a proper treatment of the nonadditivity of a microscopic free energy decomposition. Accounting for (1) helix <--> coil conformation changes along the backbone similar to the Lifson-Roig model, (2) i to i + 4 hydrogen-bond formation <--> breaking similar to the Zimm-Bragg model, and (3) structured <--> unstructured solvent interaction (hydration effects), a six-parameter DCM describes normal and inverted helix-coil transitions in polypeptides. Under suitable mixed solvent conditions heat and cold denaturation is predicted. Model parameters are fitted to experimental data showing different degrees of cold denaturation in monomeric polypeptides in aqueous hexafluoroisopropanol (HFIP) solution at various HFIP concentrations. By assuming a linear HFIP concentration dependence (up to 6% by mole fraction) on model parameters, all essential experimentally observed features are captured.
Figures








Similar articles
-
Network rigidity at finite temperature: relationships between thermodynamic stability, the nonadditivity of entropy, and cooperativity in molecular systems.Phys Rev E Stat Nonlin Soft Matter Phys. 2003 Dec;68(6 Pt 1):061109. doi: 10.1103/PhysRevE.68.061109. Epub 2003 Dec 31. Phys Rev E Stat Nonlin Soft Matter Phys. 2003. PMID: 14754182 Free PMC article.
-
Nonadditivity in the alpha-helix to coil transition.Biopolymers. 2011 Apr;95(4):240-53. doi: 10.1002/bip.21572. Epub 2010 Dec 23. Biopolymers. 2011. PMID: 21280020 Free PMC article.
-
Free energy determinants of secondary structure formation: I. alpha-Helices.J Mol Biol. 1995 Sep 22;252(3):351-65. doi: 10.1006/jmbi.1995.0502. J Mol Biol. 1995. PMID: 7563056
-
Recent advances in helix-coil theory.Biophys Chem. 2002 Dec 10;101-102:281-93. doi: 10.1016/s0301-4622(02)00170-9. Biophys Chem. 2002. PMID: 12488008 Review.
-
Hydration and heat stability effects on protein unfolding.Prog Biophys Mol Biol. 1993;59(3):237-84. doi: 10.1016/0079-6107(93)90002-2. Prog Biophys Mol Biol. 1993. PMID: 8441810 Review.
Cited by
-
Elucidating quantitative stability/flexibility relationships within thioredoxin and its fragments using a distance constraint model.J Mol Biol. 2006 May 5;358(3):882-904. doi: 10.1016/j.jmb.2006.02.015. Epub 2006 Feb 24. J Mol Biol. 2006. PMID: 16542678 Free PMC article.
-
Nonadditivity in conformational entropy upon molecular rigidification reveals a universal mechanism affecting folding cooperativity.Biophys J. 2011 Feb 16;100(4):1129-38. doi: 10.1016/j.bpj.2011.01.027. Biophys J. 2011. PMID: 21320459 Free PMC article.
-
Thermodynamic stability and flexibility characteristics of antibody fragment complexes.Protein Pept Lett. 2014;21(8):752-65. doi: 10.2174/09298665113209990051. Protein Pept Lett. 2014. PMID: 23855672 Free PMC article.
-
Conserved quantitative stability/flexibility relationships (QSFR) in an orthologous RNase H pair.Proteins. 2006 Jan 1;62(1):130-43. doi: 10.1002/prot.20745. Proteins. 2006. PMID: 16287093 Free PMC article.
-
Helix/coil nucleation: a local response to global demands.Biophys J. 2009 Dec 2;97(11):3000-9. doi: 10.1016/j.bpj.2009.09.013. Biophys J. 2009. PMID: 19948130 Free PMC article.
References
-
- Schellman JA. Compt-Rend Lab Carlsberg, Sér Chim. 1955;29(15):230–259. - PubMed
-
- Schellman JA. J Phys Chem. 1958;62:1485–1492.
-
- Kishimoto A, Mutai T, Araki K. Chem Commun (Camb) 2003;21(6):742–743. - PubMed
-
- Wallimann P, et al. J Am Chem Soc. 2003;125(5):1203–1220. - PubMed
-
- Lapidus LJ, Eaton WA, Hofricheter J. J Mol Biol. 2002;319(1):19–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

