Palindrome regeneration by template strand-switching mechanism at the origin of DNA replication of porcine circovirus via the rolling-circle melting-pot replication model
- PMID: 15308698
- PMCID: PMC506941
- DOI: 10.1128/JVI.78.17.9016-9029.2004
Palindrome regeneration by template strand-switching mechanism at the origin of DNA replication of porcine circovirus via the rolling-circle melting-pot replication model
Abstract
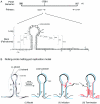
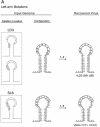
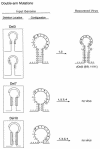
Palindromic sequences (inverted repeats) flanking the origin of DNA replication with the potential of forming single-stranded stem-loop cruciform structures have been reported to be essential for replication of the circular genomes of many prokaryotic and eukaryotic systems. In this study, mutant genomes of porcine circovirus with deletions in the origin-flanking palindrome and incapable of forming any cruciform structures invariably yielded progeny viruses containing longer and more stable palindromes. These results suggest that origin-flanking palindromes are essential for termination but not for initiation of DNA replication. Detection of template strand switching in the middle of an inverted repeat strand among the progeny viruses demonstrated that both the minus genome and a corresponding palindromic strand served as templates simultaneously during DNA biosynthesis and supports the recently proposed rolling-circle "melting-pot" replication model. The genome configuration presented by this model, a four-stranded tertiary structure, provides insights into the mechanisms of DNA replication, inverted repeat correction (or conversion), and illegitimate recombination of any circular DNA molecule with an origin-flanking palindrome.
Figures





Similar articles
-
Replication of porcine circoviruses.Virol J. 2009 May 18;6:60. doi: 10.1186/1743-422X-6-60. Virol J. 2009. PMID: 19450240 Free PMC article. Review.
-
Detection of template strand switching during initiation and termination of DNA replication of porcine circovirus.J Virol. 2004 Apr;78(8):4268-77. doi: 10.1128/jvi.78.8.4268-4277.2004. J Virol. 2004. PMID: 15047840 Free PMC article.
-
Porcine circovirus: transcription and DNA replication.Virus Res. 2012 Mar;164(1-2):46-53. doi: 10.1016/j.virusres.2011.10.012. Epub 2011 Oct 20. Virus Res. 2012. PMID: 22036834 Review.
-
Mutational analysis of the direct tandem repeat sequences at the origin of DNA replication of porcine circovirus type 1.Virology. 2005 Sep 1;339(2):192-9. doi: 10.1016/j.virol.2005.05.029. Virology. 2005. PMID: 15993915
-
A stem-loop structure, sequence non-specific, at the origin of DNA replication of porcine circovirus is essential for termination but not for initiation of rolling-circle DNA replication.Virology. 2007 Jun 20;363(1):229-35. doi: 10.1016/j.virol.2007.01.017. Epub 2007 Feb 15. Virology. 2007. PMID: 17306320
Cited by
-
The complete mitochondrial genome sequence of the green microalga Lobosphaera (Parietochloris) incisa reveals a new type of palindromic repetitive repeat.BMC Genomics. 2015 Aug 5;16(1):580. doi: 10.1186/s12864-015-1792-x. BMC Genomics. 2015. PMID: 26238519 Free PMC article.
-
Evidence of pervasive biologically functional secondary structures within the genomes of eukaryotic single-stranded DNA viruses.J Virol. 2014 Feb;88(4):1972-89. doi: 10.1128/JVI.03031-13. Epub 2013 Nov 27. J Virol. 2014. PMID: 24284329 Free PMC article.
-
Unusually long palindromes are abundant in mitochondrial control regions of insects and nematodes.PLoS One. 2006 Dec 20;1(1):e110. doi: 10.1371/journal.pone.0000110. PLoS One. 2006. PMID: 17205114 Free PMC article.
-
Mitochondrial genomes and Doubly Uniparental Inheritance: new insights from Musculista senhousia sex-linked mitochondrial DNAs (Bivalvia Mytilidae).BMC Genomics. 2011 Sep 6;12:442. doi: 10.1186/1471-2164-12-442. BMC Genomics. 2011. PMID: 21896183 Free PMC article.
-
Replication of porcine circoviruses.Virol J. 2009 May 18;6:60. doi: 10.1186/1743-422X-6-60. Virol J. 2009. PMID: 19450240 Free PMC article. Review.
References
-
- Allan, G. M., and J. A. Ellis. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Investig. 12:3-14. - PubMed
-
- Cheung, A. K. 2003. Comparative analysis of the transcriptional patterns of pathogenic and non-pathogenic circoviruses. Virology 301:41-49. - PubMed
-
- Cheung, A. K. 2004. Identification of the essential and non-essential transcriptional units for protein synthesis, DNA replication and infectious virus production of porcine circovirus type 1. Arch. Virol. 149:975-988. - PubMed
-
- Cheung, A. K. Identification of an octanucleotide motif sequence essential for viral protein, DNA and progeny virus biosynthesis at the origin of DNA replication of porcine circovirus type 2. Virology, 324:28-36. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

