Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit
- PMID: 15308704
- PMCID: PMC506919
- DOI: 10.1128/JVI.78.17.9084-9092.2004
Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit
Abstract
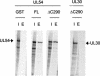
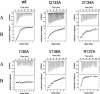
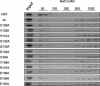
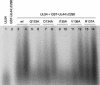
The human cytomegalovirus DNA polymerase includes an accessory protein, UL44, which has been proposed to act as a processivity factor for the catalytic subunit, UL54. How UL44 interacts with UL54 has not yet been elucidated. The crystal structure of UL44 revealed the presence of a connector loop analogous to that of the processivity subunit of herpes simplex virus DNA polymerase, UL42, which is crucial for interaction with its cognate catalytic subunit, UL30. To investigate the role of the UL44 connector loop, we replaced each of its amino acids (amino acids 129 to 140) with alanine. We then tested the effect of each substitution on the UL44-UL54 interaction by glutathione S-transferase pulldown and isothermal titration calorimetry assays, on the stimulation of UL54-mediated long-chain DNA synthesis by UL44, and on the binding of UL44 to DNA-cellulose columns. Substitutions that affected residues 133 to 136 of the connector loop measurably impaired the UL44-UL54 interaction without altering the ability of UL44 to bind DNA. One substitution, I135A, completely disrupted the binding of UL44 to UL54 and inhibited the ability of UL44 to stimulate long-chain DNA synthesis by UL54. Thus, similar to the herpes simplex virus UL30-UL42 interaction, a residue of the connector loop of the accessory subunit is crucial for UL54-UL44 interaction. However, while alteration of a polar residue of the UL42 connector loop only partially reduced binding to UL30, substitution of a hydrophobic residue of UL44 completely disrupted the UL54-UL44 interaction. This information may aid the discovery of small-molecule inhibitors of the UL44-UL54 interaction.
Figures






Similar articles
-
Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44.J Virol. 2004 Jan;78(1):158-67. doi: 10.1128/jvi.78.1.158-167.2004. J Virol. 2004. PMID: 14671097 Free PMC article.
-
Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.J Biol Chem. 2006 Feb 24;281(8):5224-32. doi: 10.1074/jbc.M506900200. Epub 2005 Dec 20. J Biol Chem. 2006. PMID: 16371349
-
Inhibition of human cytomegalovirus DNA polymerase by C-terminal peptides from the UL54 subunit.J Virol. 2003 Aug;77(15):8336-44. doi: 10.1128/jvi.77.15.8336-8344.2003. J Virol. 2003. PMID: 12857903 Free PMC article.
-
Herpesvirus DNA polymerases: Structures, functions and inhibitors.Virus Res. 2017 Apr 15;234:177-192. doi: 10.1016/j.virusres.2017.01.019. Epub 2017 Jan 30. Virus Res. 2017. PMID: 28153606 Review.
-
Clubbing together on clamps: The key to translesion synthesis.DNA Repair (Amst). 2006 Mar 7;5(3):404-7. doi: 10.1016/j.dnarep.2005.12.005. Epub 2006 Jan 19. DNA Repair (Amst). 2006. PMID: 16427367 Review.
Cited by
-
The carboxy-terminal segment of the human cytomegalovirus DNA polymerase accessory subunit UL44 is crucial for viral replication.J Virol. 2010 Nov;84(21):11563-8. doi: 10.1128/JVI.01033-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739543 Free PMC article.
-
The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner.PLoS One. 2012;7(11):e49630. doi: 10.1371/journal.pone.0049630. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166733 Free PMC article.
-
Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA.Virol J. 2005 Jul 15;2:55. doi: 10.1186/1743-422X-2-55. Virol J. 2005. PMID: 16022730 Free PMC article.
-
Role of helix P of the human cytomegalovirus DNA polymerase in resistance and hypersusceptibility to the antiviral drug foscarnet.J Virol. 2006 Feb;80(3):1440-50. doi: 10.1128/JVI.80.3.1440-1450.2006. J Virol. 2006. PMID: 16415021 Free PMC article.
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
References
-
- Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. - PubMed
-
- Bridges, K. G., Q. Hua, M. R. Brigham-Burke, J. D. Martin, P. Hensley, C. E. Dahl, P. Digard, M. A. Weiss, and D. M. Coen. 2000. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 275:472-478. - PubMed
-
- Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

