Factors that increase the effective concentration of CXCR4 dictate feline immunodeficiency virus tropism and kinetics of replication
- PMID: 15308709
- PMCID: PMC506950
- DOI: 10.1128/JVI.78.17.9132-9143.2004
Factors that increase the effective concentration of CXCR4 dictate feline immunodeficiency virus tropism and kinetics of replication
Abstract
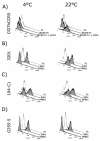
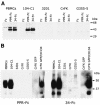
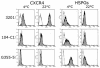
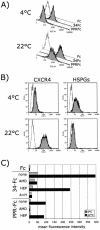
The surface glycoprotein (gp95) of the feline immunodeficiency virus (FIV) binds in a strain-specific manner to several cell surface molecules, including CXCR4, heparan sulfate proteoglycans (HSPGs), DC-SIGN, and a 43-kDa cell surface receptor on T cells recently identified as CD134 by M. Shimojima et al. (Science 303:1192-1195, 2004). CXCR4 is the entry receptor in all known cases, and the other molecules act as binding receptors to help facilitate infection. In this report, we confirm and extend the findings regarding CD134 as a primary receptor for FIV. In addition, we show that temperature critically influences the binding properties of FIV gp95 to CXCR4 and HSPGs. The data show that gp95 of the field strain FIV-PPR bound to CXCR4 at 22 degrees C, whereas binding was not detected at 4 degrees C. In contrast, binding of the laboratory adapted FIV-34TF10 gp95 was observed at either 4 degrees C or 22 degrees C, albeit at increased levels at the higher temperature. The level of CXCR4 increased after the temperature was switched from 4 to 22 degrees C, whereas the level of HSPGs decreased, resulting in higher binding of gp95 from both strains to CXCR4 and lower binding of gp95 of FIV-34TF10 to HSPGs (FIV-PPR gp95 does not bind to these molecules). The findings also show that HSPGs facilitate the CXCR4-mediated infectivity of CrFK and G355-5 cells by FIV-34TF10. These two nonlymphoid cell lines express very low levels of CXCR4 and are permissive to FIV-34TF10 but not to productive infection by FIV-PPR. However, overexpression of human CXCR4 in CrFK or G-355-5 cells resulted in extensive cell fusion and infection by FIV-PPR. Taken together, these findings indicate that factors that increase the effective concentration of CXCR4 enhance FIV infectivity and may involve (i) temperature or ligand-induced conformational changes in CXCR4 that enhance SU binding, (ii) coreceptor interactions with gp95 that either alter gp95 conformation to enhance CXCR4 binding and/or raise the localized concentration of receptor or ligand, or (iii) direct increase in CXCR4 concentration via overexpression.
Figures





Similar articles
-
Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor.J Virol. 2001 May;75(10):4528-39. doi: 10.1128/JVI.75.10.4528-4539.2001. J Virol. 2001. PMID: 11312323 Free PMC article.
-
Identification of amino acid residues important for heparan sulfate proteoglycan interaction within variable region 3 of the feline immunodeficiency virus surface glycoprotein.J Virol. 2011 Jul;85(14):7108-17. doi: 10.1128/JVI.00573-11. Epub 2011 May 4. J Virol. 2011. PMID: 21543468 Free PMC article.
-
CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells.J Virol. 1998 Aug;72(8):6858-66. doi: 10.1128/JVI.72.8.6858-6866.1998. J Virol. 1998. PMID: 9658135 Free PMC article.
-
The virus-receptor interaction in the replication of feline immunodeficiency virus (FIV).Curr Opin Virol. 2013 Dec;3(6):670-5. doi: 10.1016/j.coviro.2013.08.003. Epub 2013 Aug 28. Curr Opin Virol. 2013. PMID: 23992667 Free PMC article. Review.
-
[Feline immunodeficiency virus tropism].Uirusu. 2007 Jun;57(1):75-82. doi: 10.2222/jsv.57.75. Uirusu. 2007. PMID: 18040157 Review. Japanese.
Cited by
-
Thymic pathogenicity of an HIV-1 envelope is associated with increased CXCR4 binding efficiency and V5-gp41-dependent activity, but not V1/V2-associated CD4 binding efficiency and viral entry.Virology. 2005 Jun 5;336(2):184-97. doi: 10.1016/j.virol.2005.03.032. Virology. 2005. PMID: 15892960 Free PMC article.
-
Fine definition of the CXCR4-binding region on the V3 loop of feline immunodeficiency virus surface glycoprotein.PLoS One. 2010 May 18;5(5):e10689. doi: 10.1371/journal.pone.0010689. PLoS One. 2010. PMID: 20502526 Free PMC article.
-
Mapping of the CXCR4 binding site within variable region 3 of the feline immunodeficiency virus surface glycoprotein.J Virol. 2008 Sep;82(18):9134-42. doi: 10.1128/JVI.00394-08. Epub 2008 Jul 2. J Virol. 2008. PMID: 18596086 Free PMC article.
-
Mutations in the feline immunodeficiency virus envelope glycoprotein confer resistance to a dominant-negative fragment of Tsg101 by enhancing infectivity and cell-to-cell virus transmission.Biochim Biophys Acta. 2014 Apr;1838(4):1143-52. doi: 10.1016/j.bbamem.2013.08.020. Epub 2013 Sep 10. Biochim Biophys Acta. 2014. PMID: 24036228 Free PMC article.
-
Vaccine-associated enhanced disease in humans and animal models: Lessons and challenges for vaccine development.Front Microbiol. 2022 Aug 10;13:932408. doi: 10.3389/fmicb.2022.932408. eCollection 2022. Front Microbiol. 2022. PMID: 36033843 Free PMC article. Review.
References
-
- Babcock, G. J., T. Mirzabekov, W. Wojtowicz, and J. Sodroski. 2001. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 276:38433-38440. - PubMed
-
- Bobardt, M. D., A. C. Saphire, H. C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. A. Gallay. 2003. Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 18:27-39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

