The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex
- PMID: 15308710
- PMCID: PMC506948
- DOI: 10.1128/JVI.78.17.9144-9153.2004
The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex
Abstract
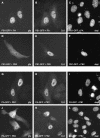
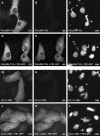
The RNA genome of influenza virus is transcribed and replicated by the viral RNA polymerase complex in the cell nucleus. We have generated green fluorescent protein (GFP)-tagged polymerase subunits to study the assembly of the polymerase complex. Our results show that individually expressed polymerase basic protein 1 (PB1) and polymerase acidic protein (PA) subunits were distributed in both the cytoplasm and the nucleus, while the polymerase basic protein 2 (PB2) subunit accumulated in the nucleus. Although it has been reported that PB1 alone accumulates in the nucleus, we demonstrate that PB1 requires the coexpression of PA for efficient nuclear accumulation. Our results support a model which proposes that PB1 and PA are transported into the nucleus as a complex.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

