Kinetic analysis of the interactions between vaccinia virus complement control protein and human complement proteins C3b and C4b
- PMID: 15308738
- PMCID: PMC506936
- DOI: 10.1128/JVI.78.17.9446-9457.2004
Kinetic analysis of the interactions between vaccinia virus complement control protein and human complement proteins C3b and C4b
Abstract
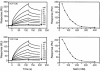
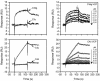
The vaccinia virus complement control protein (VCP) is an immune evasion protein of vaccinia virus. Previously, VCP has been shown to bind and support inactivation of host complement proteins C3b and C4b and to protect the vaccinia virions from antibody-dependent complement-enhanced neutralization. However, the molecular mechanisms involved in the interaction of VCP with its target proteins C3b and C4b have not yet been elucidated. We have utilized surface plasmon resonance technology to study the interaction of VCP with C3b and C4b. We measured the kinetics of binding of the viral protein to its target proteins and compared it with human complement regulators factor H and sCR1, assessed the influence of immobilization of ligand on the binding kinetics, examined the effect of ionic contacts on these interactions, and sublocalized the binding site on C3b and C4b. Our results indicate that (i) the orientation of the ligand is important for accurate determination of the binding constants, as well as the mechanism of binding; (ii) in contrast to factor H and sCR1, the binding of VCP to C3b and C4b follows a simple 1:1 binding model and does not involve multiple-site interactions as predicted earlier; (iii) VCP has a 4.6-fold higher affinity for C4b than that for C3b, which is also reflected in its factor I cofactor activity; (iv) ionic interactions are important for VCP-C3b and VCP-C4b complex formation; (v) VCP does not bind simultaneously to C3b and C4b; and (vi) the binding site of VCP on C3b and C4b is located in the C3dg and C4c regions, respectively.
Figures

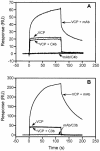
 , Factor H binds to C4b only in a buffer with very low ionic strength (47).
, Factor H binds to C4b only in a buffer with very low ionic strength (47).




References
-
- Ahearn, J. M., M. B. Fischer, D. Croix, S. Goerg, M. Ma, J. Xia, X. Zhou, R. G. Howard, T. L. Rothstein, and M. C. Carroll. 1996. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B-cell response to T-dependent antigen. Immunity 4:251-262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

