Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates
- PMID: 15308750
- PMCID: PMC506943
- DOI: 10.1128/JVI.78.17.9568-9572.2004
Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates
Abstract
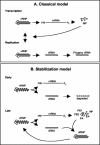
The RNA-dependent RNA polymerase of influenza A virus is responsible for both transcription and replication of negative-sense viral RNA. It is thought that a "switching" mechanism regulates the transition between these activities. We demonstrate that, in the presence of preexisting viral RNA polymerase and nucleoprotein (NP), influenza A virus synthesizes both mRNA (transcription) and cRNA (replication) early in infection. We suggest that there may be no switch regulating the initiation of RNA synthesis and present a model suggesting that nascent cRNA is degraded by host cell nucleases unless it is stabilized by newly synthesized viral RNA polymerase and NP.
Figures




References
-
- Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 2002. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim. Biophys. Acta 1577:337-353. - PubMed
-
- Barrett, T., A. J. Wolstenholme, and B. W. J. Mahy. 1979. Transcription and replication of influenza virus RNA. Virology 98:211-225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

