Characterization of the human cytomegalovirus UL34 gene
- PMID: 15308752
- PMCID: PMC506942
- DOI: 10.1128/JVI.78.17.9579-9583.2004
Characterization of the human cytomegalovirus UL34 gene
Abstract
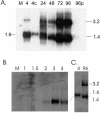
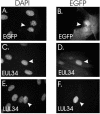
UL34 encodes the transcriptional repressor of the human cytomegalovirus immune evasion gene, US3, and is essential for viral replication in tissue culture. Two different monocistronic transcripts originate from UL34 at early and late times postinfection and encode two predominant proteins and a third, minor protein. The UL34 proteins are differentially expressed throughout the viral replication cycle, with both proteins localizing to the nucleus and repressing expression of the US3 gene.
Figures


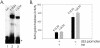
Similar articles
-
Identification of the functional domains of the essential human cytomegalovirus UL34 proteins.Virology. 2006 Sep 15;353(1):27-34. doi: 10.1016/j.virol.2006.05.019. Epub 2006 Jun 19. Virology. 2006. PMID: 16784766
-
Human cytomegalovirus UL34 early and late proteins are essential for viral replication.Viruses. 2014 Jan 28;6(2):476-88. doi: 10.3390/v6020476. Viruses. 2014. PMID: 24476753 Free PMC article.
-
Primate cytomegaloviruses encode and express an IL-10-like protein.Virology. 2000 Mar 15;268(2):272-80. doi: 10.1006/viro.2000.0195. Virology. 2000. PMID: 10704336
-
The proteins of human cytomegalovirus.Birth Defects Orig Artic Ser. 1984;20(1):49-62. Birth Defects Orig Artic Ser. 1984. PMID: 6329373 Review. No abstract available.
-
Human cytomegalovirus: host immune modulation by the viral US3 gene.Int J Biochem Cell Biol. 2009 Mar;41(3):503-6. doi: 10.1016/j.biocel.2008.10.012. Epub 2008 Oct 18. Int J Biochem Cell Biol. 2009. PMID: 18992841 Review.
Cited by
-
Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface.PLoS Pathog. 2013;9(9):e1003611. doi: 10.1371/journal.ppat.1003611. Epub 2013 Sep 26. PLoS Pathog. 2013. PMID: 24086132 Free PMC article.
-
Decoding human cytomegalovirus.Science. 2012 Nov 23;338(6110):1088-93. doi: 10.1126/science.1227919. Science. 2012. PMID: 23180859 Free PMC article.
-
pUL34 binding near the human cytomegalovirus origin of lytic replication enhances DNA replication and viral growth.Virology. 2018 May;518:414-422. doi: 10.1016/j.virol.2018.03.017. Epub 2018 Apr 5. Virology. 2018. PMID: 29626748 Free PMC article.
-
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.Viruses. 2023 Aug 8;15(8):1703. doi: 10.3390/v15081703. Viruses. 2023. PMID: 37632045 Free PMC article. Review.
-
UL34 Deletion Restricts Human Cytomegalovirus Capsid Formation and Maturation.Int J Mol Sci. 2022 May 21;23(10):5773. doi: 10.3390/ijms23105773. Int J Mol Sci. 2022. PMID: 35628580 Free PMC article.
References
-
- Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

