Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY
- PMID: 15308775
- PMCID: PMC515128
- DOI: 10.1073/pnas.0404287101
Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY
Abstract
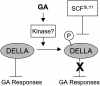
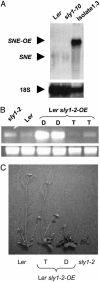
This article reports the genetic interaction of two F-box genes, SLEEPY1 (SLY1) and SNEEZY (SNE), in Arabidopsis thaliana gibberellin (GA) signaling. The SLY1 gene encodes an F-box subunit of a Skp1-cullin-F-box (SCF) E3 ubiquitin ligase complex that positively regulates GA signaling. The sly1-2 and sly1-10 mutants have recessive, GA-insensitive phenotypes including delayed germination, dwarfism, reduced fertility, and overaccumulation of the DELLA proteins RGA (Repressor of ga1-3), GAI (GA-Insensitive), and RGL2 (RGA-Like 2). The DELLA domain proteins are putative transcription factors that negatively regulate GA signaling. The requirement for SLY1 in GA-stimulated disappearance of DELLA proteins suggests that GA targets DELLA proteins for destruction via SCF(SLY1)-mediated ubiquitylation. Overexpression of SLY1 in sly1-2 and sly1-10 plants rescues the recessive GA-insensitive phenotype of these mutants. Surprisingly, antisense expression of SLY1 also suppresses these mutants. This result caused us to hypothesize that the SLY1 homologue SNE can functionally replace SLY1 in the absence of the recessive interfering sly1-2 or sly1-10 genes. This hypothesis was supported because overexpression of SNE in sly1-10 rescues the dwarf phenotype. In addition to rescuing the sly1-10 dwarf phenotype, SNE overexpression also restored normal RGA protein levels, suggesting that the SNE F-box protein can replace SLY1 in the GA-induced proteolysis of RGA. If the C-terminal truncation in the sly1-2 and sly1-10 alleles interferes with SNE rescue, we reasoned that overexpression of sly1-2 might interfere with wild-type SLY1 function. Indeed, overexpression of sly1-2 in wild-type Ler (Landsberg erecta) yields dwarf plants.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

