Alternative mapping of probes to genes for Affymetrix chips
- PMID: 15310390
- PMCID: PMC514699
- DOI: 10.1186/1471-2105-5-111
Alternative mapping of probes to genes for Affymetrix chips
Abstract
Background: Short oligonucleotide arrays have several probes measuring the expression level of each target transcript. Therefore the selection of probes is a key component for the quality of measurements. However, once probes have been selected and synthesized on an array, it is still possible to re-evaluate the results using an updated mapping of probes to genes, taking into account the latest biological knowledge available.
Methods: We investigated how probes found on recent commercial microarrays for human genes (Affymetrix HG-U133A) were matching a recent curated collection of human transcripts: the NCBI RefSeq database. We also built mappings and used them in place of the original probe to genes associations provided by the manufacturer of the arrays.
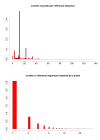
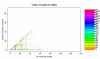
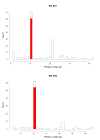
Results: In a large number of cases, 36%, the probes matching a reference sequence were consistent with the grouping of probes by the manufacturer of the chips. For the remaining cases there were discrepancies and we show how that can affect the analysis of data.
Conclusions: While the probes on Affymetrix arrays remain the same for several years, the biological knowledge concerning the genomic sequences evolves rapidly. Using up-to-date knowledge can apparently change the outcome of an analysis.
Figures

References
-
- Affymetrix Affymetrix Microarray Suite User Guide. version 4, Affymetrix, Santa Clara, CA. 1999.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

