Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells
- PMID: 15310660
- PMCID: PMC515319
- DOI: 10.1101/gr.2431504
Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells
Abstract
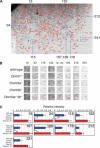
Many CpG islands have tissue-dependent and differentially methylated regions (T-DMRs) in normal cells and tissues. To elucidate how DNA methyltransferases (Dnmts) participate in methylation of the genomic components, we investigated the genome-wide DNA methylation pattern of the T-DMRs with Dnmt1-, Dnmt3a-, and/or Dnmt3b-deficient ES cells by restriction landmark genomic scanning (RLGS). Approximately 1300 spots were detected in wild-type ES cells. In Dnmt1(-/-) ES cells, additional 236 spots emerged, indicating that the corresponding loci are methylated by Dnmt1 in wild-type ES cells. Intriguingly, in Dnmt3a(-/-)Dnmt3b(-/-) ES cells, the same 236 spots also emerged, and no additional spots appeared differentially. Therefore, Dnmt1 and Dnmt3a/3b share targets in CpG islands. Cloning and virtual image RLGS revealed that 81% of the RLGS spots were associated with genes, and 62% of the loci were in CpG islands. By contrast to the previous reports that demethylation at repeated sequences was severe in Dnmt1(-/-) cells compared with Dnmt3a(-/-)Dnmt3b(-/-) cells, a complete loss of methylation was observed at RLGS loci in Dnmt3a(-/-)Dnmt3b(-/-) cells, whereas methylation levels only decreased to 16% to 48% in the Dnmt1(-/-) cells. We concluded that there are CpG islands with T-DMR as targets shared by Dnmt1 and Dnmt3a/3b and that each Dnmt has target preferences depending on the genomic components.
Figures
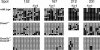


Similar articles
-
Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b.Genes Cells. 2006 Jul;11(7):805-14. doi: 10.1111/j.1365-2443.2006.00984.x. Genes Cells. 2006. PMID: 16824199
-
Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells.Genes Cells. 2007 Jan;12(1):1-11. doi: 10.1111/j.1365-2443.2006.01029.x. Genes Cells. 2007. PMID: 17212651
-
Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation.Genes Cells. 2006 Oct;11(10):1225-37. doi: 10.1111/j.1365-2443.2006.01012.x. Genes Cells. 2006. PMID: 16999741
-
Dynamic regulation of DNA methylation during mammalian development.Epigenomics. 2009 Oct;1(1):81-98. doi: 10.2217/epi.09.5. Epigenomics. 2009. PMID: 22122638 Review.
-
The study of aberrant methylation in cancer via restriction landmark genomic scanning.Oncogene. 2002 Aug 12;21(35):5414-26. doi: 10.1038/sj.onc.1205608. Oncogene. 2002. PMID: 12154404 Review.
Cited by
-
Establishment of trophoblast stem cell lines from somatic cell nuclear-transferred embryos.Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16293-7. doi: 10.1073/pnas.0908009106. Epub 2009 Aug 24. Proc Natl Acad Sci U S A. 2009. PMID: 19706390 Free PMC article.
-
DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner.Genes Dev. 2006 Dec 15;20(24):3382-94. doi: 10.1101/gad.1470906. Genes Dev. 2006. PMID: 17182866 Free PMC article.
-
Stable transmission of reversible modifications: maintenance of epigenetic information through the cell cycle.Cell Mol Life Sci. 2011 Jan;68(1):27-44. doi: 10.1007/s00018-010-0505-5. Epub 2010 Aug 27. Cell Mol Life Sci. 2011. PMID: 20799050 Free PMC article. Review.
-
DNA methylation is required for the control of stem cell differentiation in the small intestine.Genes Dev. 2014 Mar 15;28(6):652-64. doi: 10.1101/gad.230318.113. Genes Dev. 2014. PMID: 24637118 Free PMC article.
-
DNA methylation in hematopoietic development and disease.Exp Hematol. 2016 Sep;44(9):783-790. doi: 10.1016/j.exphem.2016.04.013. Epub 2016 May 10. Exp Hematol. 2016. PMID: 27178734 Free PMC article. Review.
References
-
- Bird, A.P. 1987. CpG island as gene markers in the vertebrate nucleus. Trends Genet. 3: 342-347.
-
- Bird, A.P. and Wolffe, A.P. 1999. Methylation-induced repression: Belts, braces, and chromatin. Cell 99: 451-454. - PubMed
WEB SITE REFERENCES
-
- http://www.ensembl.org/Mus_musculus/; Ensemble.
-
- ftp://ftp.ncbi.nih.gov/genomes/M_musculus/; GenBank ftp site.
-
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
