Regional glucose metabolism and glutamatergic neurotransmission in rat brain in vivo
- PMID: 15310848
- PMCID: PMC515118
- DOI: 10.1073/pnas.0405065101
Regional glucose metabolism and glutamatergic neurotransmission in rat brain in vivo
Abstract
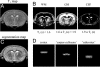
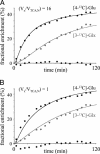
Multivolume (1)H-[(13)C] NMR spectroscopy in combination with i.v. [1,6-(13)C(2)]glucose infusion was used to detect regional glucose metabolism and glutamatergic neurotransmission in the halothane-anesthetized rat brain at 7 T. The regional information was decomposed into pure cerebral gray matter, white matter, and subcortical structures by means of tissue segmentation based on quantitative T(1) relaxation mapping. The (13)C turnover curves of [4-(13)C]glutamate, [4-(13)C]glutamine, and [3-(13)C]glutamate + glutamine were fitted with a two-compartment neuronal-astroglial metabolic model. The neuronal tricarboxylic acid cycle fluxes in cerebral gray matter, white matter, and subcortex were 0.79 +/- 0.15, 0.20 +/- 0.11, and 0.42 +/- 0.09 micromol/min per g, respectively. The glutamate-glutamine neurotransmitter cycle fluxes in cerebral gray matter, white matter, and subcortex were 0.31 +/- 0.07, 0.02 +/- 0.04, and 0.18 +/- 0.12 micromol/min per g, respectively. The exchange rate between the mitochondrial and cytosolic metabolite pools was fast relative to the neuronal tricarboxylic acid cycle flux for all cerebral tissue types.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

