Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases
- PMID: 15310852
- PMCID: PMC515083
- DOI: 10.1073/pnas.0403454101
Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases
Abstract
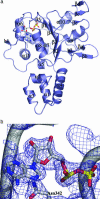
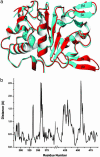
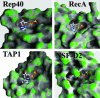
We have determined the structure of adeno-associated virus type 2 (AAV2) Rep40 to 2.1-A resolution with ADP bound at the active site. The complex crystallizes as a monomer with one ADP molecule positioned in an unexpectedly open binding site. The nucleotide-binding pocket consists of the P-loop residues interacting with the phosphates and a loop (nucleoside-binding loop) that emanates from the last strand of the central beta-sheet and interacts with the sugar and base. As a result of the open nature of the binding site, one face of the adenine ring is completely exposed to the solvent, and consequently the number of protein-nucleotide contacts is scarce as compared with other P-loop nucleotide phosphohydrolases. The conformation of the ADP molecule in its binding site bears a resemblance to those found in only three other families of P-loop ATPases: the ATP-binding cassette transporter family, the bacterial RecA proteins, and the type II topoisomerase family. In all these cases, oligomerization is required to attain a competent nucleotide-binding pocket. We propose that this characteristic is native to superfamily 3 helicases and allows for an additional mechanism of regulation by these multifunctional proteins. Furthermore, it explains the strong tendency by members of this family such as simian virus 40 TAg to oligomerize after binding ATP.
Figures

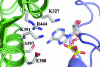
Similar articles
-
Crystal structure of the SF3 helicase from adeno-associated virus type 2.Structure. 2003 Aug;11(8):1025-35. doi: 10.1016/s0969-2126(03)00152-7. Structure. 2003. PMID: 12906833
-
Crystal structure of a DExx box DNA helicase.Nature. 1996 Nov 28;384(6607):379-83. doi: 10.1038/384379a0. Nature. 1996. PMID: 8934527
-
The 1.5 A resolution crystal structure of the carbamate kinase-like carbamoyl phosphate synthetase from the hyperthermophilic Archaeon pyrococcus furiosus, bound to ADP, confirms that this thermostable enzyme is a carbamate kinase, and provides insight into substrate binding and stability in carbamate kinases.J Mol Biol. 2000 Jun 2;299(2):463-76. doi: 10.1006/jmbi.2000.3779. J Mol Biol. 2000. PMID: 10860751
-
Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA.Science. 2002 Sep 20;297(5589):2018-26. doi: 10.1126/science.1074424. Science. 2002. PMID: 12242434
-
Modularity and specialization in superfamily 1 and 2 helicases.J Bacteriol. 2002 Apr;184(7):1819-26. doi: 10.1128/JB.184.7.1819-1826.2002. J Bacteriol. 2002. PMID: 11889086 Free PMC article. Review. No abstract available.
Cited by
-
DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68.PLoS Pathog. 2009 Jul;5(7):e1000513. doi: 10.1371/journal.ppat.1000513. Epub 2009 Jul 10. PLoS Pathog. 2009. PMID: 19593381 Free PMC article.
-
Evidence for a structural relationship between BRCT domains and the helicase domains of the replication initiators encoded by the Polyomaviridae and Papillomaviridae families of DNA tumor viruses.J Virol. 2008 Sep;82(17):8849-62. doi: 10.1128/JVI.00553-08. Epub 2008 Jun 25. J Virol. 2008. PMID: 18579587 Free PMC article.
-
Mutations in Sensor 1 and Walker B in the bovine papillomavirus E1 initiator protein mimic the nucleotide-bound state.J Virol. 2010 Feb;84(4):1912-9. doi: 10.1128/JVI.01756-09. Epub 2009 Nov 25. J Virol. 2010. PMID: 19939914 Free PMC article.
-
Characterization of papillomavirus E1 helicase mutants defective for interaction with the SUMO-conjugating enzyme Ubc9.Virology. 2009 Dec 20;395(2):190-201. doi: 10.1016/j.virol.2009.09.020. Virology. 2009. PMID: 19836047 Free PMC article.
-
Functional and evolutionary analysis of viral proteins containing a Rossmann-like fold.Protein Sci. 2018 Aug;27(8):1450-1463. doi: 10.1002/pro.3438. Epub 2018 Jun 13. Protein Sci. 2018. PMID: 29722076 Free PMC article.
References
-
- Samulski, R. J. (2003) Ernst Schering Res. Found. Workshop 43, 25–40. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

