Kinase-independent transcriptional co-activation of peroxisome proliferator-activated receptor alpha by AMP-activated protein kinase
- PMID: 15312046
- PMCID: PMC1134113
- DOI: 10.1042/BJ20040955
Kinase-independent transcriptional co-activation of peroxisome proliferator-activated receptor alpha by AMP-activated protein kinase
Abstract
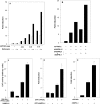
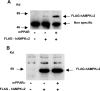
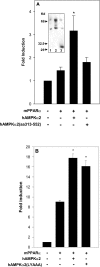
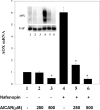
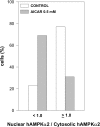
AMPK (AMP-activated protein kinase) responds to intracellular ATP depletion, while PPARalpha (peroxisome proliferator-activated receptor alpha) induces the expression of genes coding for enzymes and proteins involved in increasing cellular ATP yields. PPARalpha-mediated transcription is shown here to be co-activated by the alpha subunit of AMPK, as well as by kinase-deficient (Thr172Ala) and kinase-less (Asp157Ala, Asp139Ala) mutants of AMPKalpha. The Ser452Ala mutant of mPPARalpha mutated in its putative consensus AMPKalpha phosphorylation site is similarly co-activated by AMPKalpha. AMPKalpha or its kinase-less mutants bind to PPARalpha; binding is increased by MgATP, to a lesser extent by MgADP, but not at all by AMP or ZMP [AICAR (5-aminoimidazole-4-carboxamide ribonucleoside) monophosphate]. ATP-activated binding of AMPKalpha to PPARalpha is mediated primarily by the C-terminal regulatory domain of AMPKalpha. PPARalpha co-activation by AMPKalpha may, however, require its secondary interaction with the N-terminal catalytic domain of AMPKalpha, independently of its kinase activity. While AMPK catalytic activity is activated by AICAR, PPARalpha co-activation and PPARalpha-controlled transcription are robustly inhibited by AICAR, with concomitant translocation of nuclear AMPKalpha or its kinase-less mutants to the cytosol. In conclusion, AMPKalpha, independently of its kinase activity, co-activates PPARalpha both in primary rat hepatocytes and in PPARalpha-transfected cells. The kinase and transcriptional co-activation modes of AMPKalpha are both regulated by the cellular ATP/AMP ratio. Co-activation of PPARalpha by AMPKalpha may transcriptionally complement AMPK in maintaining cellular ATP status.
Figures
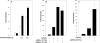

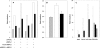

Similar articles
-
Stimulation of hepatocytic AMP-activated protein kinase by okadaic acid and other autophagy-suppressive toxins.Biochem J. 2005 Mar 1;386(Pt 2):237-44. doi: 10.1042/BJ20040609. Biochem J. 2005. PMID: 15461583 Free PMC article.
-
AMPK activation enhances PPARα activity to inhibit cardiac hypertrophy via ERK1/2 MAPK signaling pathway.Arch Biochem Biophys. 2011 Jul;511(1-2):1-7. doi: 10.1016/j.abb.2011.04.010. Epub 2011 Apr 21. Arch Biochem Biophys. 2011. PMID: 21530483
-
AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARalpha and PGC-1.Biochem Biophys Res Commun. 2006 Feb 3;340(1):291-5. doi: 10.1016/j.bbrc.2005.12.011. Epub 2005 Dec 12. Biochem Biophys Res Commun. 2006. PMID: 16364253
-
AMP-activated protein kinase in metabolic control and insulin signaling.Circ Res. 2007 Feb 16;100(3):328-41. doi: 10.1161/01.RES.0000256090.42690.05. Circ Res. 2007. PMID: 17307971 Review.
-
Selective suppression of AMP-activated protein kinase in skeletal muscle: update on 'lazy mice'.Biochem Soc Trans. 2003 Feb;31(Pt 1):236-41. doi: 10.1042/bst0310236. Biochem Soc Trans. 2003. PMID: 12546693 Review.
Cited by
-
Tang-Nai-Kang alleviates pre-diabetes and metabolic disorders and induces a gene expression switch toward fatty acid oxidation in SHR.Cg-Leprcp/NDmcr rats.PLoS One. 2015 Apr 13;10(4):e0122024. doi: 10.1371/journal.pone.0122024. eCollection 2015. PLoS One. 2015. PMID: 25874615 Free PMC article.
-
Healthy Effects of Plant Polyphenols: Molecular Mechanisms.Int J Mol Sci. 2020 Feb 13;21(4):1250. doi: 10.3390/ijms21041250. Int J Mol Sci. 2020. PMID: 32070025 Free PMC article. Review.
-
Diospyros kaki and Citrus unshiu Mixture Improves Disorders of Lipid Metabolism in Nonalcoholic Fatty Liver Disease.Can J Gastroenterol Hepatol. 2020 Dec 24;2020:8812634. doi: 10.1155/2020/8812634. eCollection 2020. Can J Gastroenterol Hepatol. 2020. PMID: 33425805 Free PMC article.
-
Erythropoietin contributes to slow oxidative muscle fiber specification via PGC-1α and AMPK activation.Int J Biochem Cell Biol. 2013 Jul;45(7):1155-64. doi: 10.1016/j.biocel.2013.03.007. Epub 2013 Mar 20. Int J Biochem Cell Biol. 2013. PMID: 23523698 Free PMC article.
-
Endurance factors improve hippocampal neurogenesis and spatial memory in mice.Learn Mem. 2011 Jan 18;18(2):103-7. doi: 10.1101/lm.2001611. Print 2011 Feb. Learn Mem. 2011. PMID: 21245211 Free PMC article.
References
-
- Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. - PubMed
-
- Latruffe N., Malki M. C., Nicolas-Frances V., Clemencet M. C., Jannin B., Berlot J. P. Regulation of the peroxisomal beta-oxidation-dependent pathway by peroxisome proliferator-activated receptor alpha and kinases. Biochem. Pharmacol. 2000;60:1027–1032. - PubMed
-
- Hardie D. G., Carling D., Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. - PubMed
-
- Kemp B. E., Mitchelhill K. I., Stapleton D., Michell B. J., Chen Z. P., Witters L. A. Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem. Sci. 1999;24:22–25. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

