The B-subunit of DNA polymerase alpha-primase associates with the origin recognition complex for initiation of DNA replication
- PMID: 15314153
- PMCID: PMC506996
- DOI: 10.1128/MCB.24.17.7419-7434.2004
The B-subunit of DNA polymerase alpha-primase associates with the origin recognition complex for initiation of DNA replication
Abstract
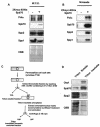
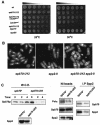
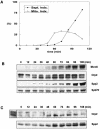
The B-subunit (p70/Pol12p) of the DNA polymerase alpha-primase (Polalpha-primase) complex is thought to have a regulatory role in an early stage of S phase. We generated a panel of fission yeast thermosensitive mutants of the B-subunit (termed Spb70) to investigate its role in initiation of DNA replication by genetic and biochemical approaches. Here, we show that the fission yeast Spb70 genetically interacts and coprecipitates with origin recognition complex proteins Orp1/Orc1 and Orp2/Orc2 and primase coupling subunit Spp2/p58. A fraction of Spb70 associates with Orp2 on chromatin throughout the cell cycle independent of the other subunits of Polalpha-primase. Furthermore, primase Spp2/p58 subunit preferentially associates with the unphosphorylated Orp2, and the association requires Spb70. Mutations in orp2+ that abolish or mimic the Cdc2 phosphorylation of Orp2 suppress or exacerbate the thermosensitivity of the spb70 mutants, respectively, indicating that an unphosphorylated Orp2 promotes an Spb70-dependent replication event. Together, these results indicate that the chromatin-bound B-subunit in association with origin recognition complex mediates recruiting Polalpha-primase complex onto replication origins in G1 pre-Start through an interaction with primase Spp2/p58 subunit. Our results thus suggest a role for the recruited Polalpha-primase in the initiation of both leading and lagging strands at the replication origins.
Copyright 2004 American Society for Microbiology
Figures





Similar articles
-
The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex.J Biol Chem. 2014 Aug 8;289(32):22021-34. doi: 10.1074/jbc.M114.570333. Epub 2014 Jun 24. J Biol Chem. 2014. PMID: 24962573 Free PMC article.
-
Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha.Mol Cell. 2004 Oct 22;16(2):173-85. doi: 10.1016/j.molcel.2004.09.017. Mol Cell. 2004. PMID: 15494305
-
Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polα/Primase/Ctf4 Complex.Mol Cell. 2015 Mar 5;57(5):812-823. doi: 10.1016/j.molcel.2014.12.038. Epub 2015 Feb 5. Mol Cell. 2015. PMID: 25661486 Free PMC article.
-
Origins and complexes: the initiation of DNA replication.J Exp Bot. 2001 Feb;52(355):193-202. J Exp Bot. 2001. PMID: 11283163 Review.
-
New Insights into the Mechanism of DNA Duplication by the Eukaryotic Replisome.Trends Biochem Sci. 2016 Oct;41(10):859-871. doi: 10.1016/j.tibs.2016.07.011. Epub 2016 Aug 20. Trends Biochem Sci. 2016. PMID: 27555051 Review.
Cited by
-
Flexibility and distributive synthesis regulate RNA priming and handoff in human DNA polymerase α-primase.bioRxiv [Preprint]. 2023 Aug 1:2023.08.01.551538. doi: 10.1101/2023.08.01.551538. bioRxiv. 2023. Update in: J Mol Biol. 2023 Dec 15;435(24):168330. doi: 10.1016/j.jmb.2023.168330. PMID: 37577606 Free PMC article. Updated. Preprint.
-
DNA polymerases at the eukaryotic fork-20 years later.Mutat Res. 2010 Mar 1;685(1-2):45-53. doi: 10.1016/j.mrfmmm.2009.08.002. Epub 2009 Aug 12. Mutat Res. 2010. PMID: 19682465 Free PMC article. Review.
-
Crystal Structure of the Human Pol α B Subunit in Complex with the C-terminal Domain of the Catalytic Subunit.J Biol Chem. 2015 Jun 5;290(23):14328-37. doi: 10.1074/jbc.M115.649954. Epub 2015 Apr 6. J Biol Chem. 2015. PMID: 25847248 Free PMC article.
-
Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast.BMC Mol Biol. 2005 Jun 7;6:13. doi: 10.1186/1471-2199-6-13. BMC Mol Biol. 2005. PMID: 15941470 Free PMC article.
-
DNA replication: Mechanisms and therapeutic interventions for diseases.MedComm (2020). 2023 Feb 5;4(1):e210. doi: 10.1002/mco2.210. eCollection 2023 Feb. MedComm (2020). 2023. PMID: 36776764 Free PMC article. Review.
References
-
- Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
-
- Arezi, B., and R. D. Kuchta. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25:572-576. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
