SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans
- PMID: 15314158
- PMCID: PMC506987
- DOI: 10.1128/MCB.24.17.7483-7490.2004
SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans
Abstract
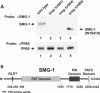
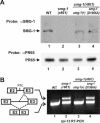
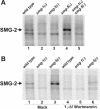
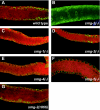
Eukaryotic messenger RNAs containing premature stop codons are selectively and rapidly degraded, a phenomenon termed nonsense-mediated mRNA decay (NMD). Previous studies with both Caenohabditis elegans and mammalian cells indicate that SMG-2/human UPF1, a central regulator of NMD, is phosphorylated in an SMG-1-dependent manner. We report here that smg-1, which is required for NMD in C. elegans, encodes a protein kinase of the phosphatidylinositol kinase superfamily of protein kinases. We identify null alleles of smg-1 and demonstrate that SMG-1 kinase activity is required in vivo for NMD and in vitro for SMG-2 phosphorylation. SMG-1 and SMG-2 coimmunoprecipitate from crude extracts, and this interaction is maintained in smg-3 and smg-4 mutants, both of which are required for SMG-2 phosphorylation in vivo and in vitro. SMG-2 is located diffusely through the cytoplasm, and its location is unaltered in mutants that disrupt the cycle of SMG-2 phosphorylation. We discuss the role of SMG-2 phosphorylation in NMD.
Copyright 2004 American Society for Microbiology
Figures


References
-
- Aronoff, R., R. Baran, and J. Hodgkin. 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene 268:153-164. - PubMed
-
- Bar-Peled, M., and N. V. Raikhel. 1996. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241:140-142. - PubMed
-
- Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
