Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability
- PMID: 15314207
- PMCID: PMC514376
- DOI: 10.1093/nar/gkh754
Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability
Abstract
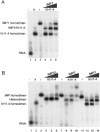
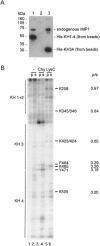
Active cytoplasmic RNA localization depends on the attachment of RNA-binding proteins that dictate the destination of the RNA molecule. In this study, we used an electrophoretic mobility-shift assay in combination with equilibrium and kinetic analyses to characterize the assembly of the human zipcode-binding protein IMP1 on targets in the 3'-UTR from Igf-II mRNA and in H19 RNA. In both cases, two molecules of IMP1 bound to RNA by a sequential, cooperative mechanism, characterized by an initial fast step, followed by a slow second step. The first step created an obligatory assembly intermediate of low stability, whereas the second step was the discriminatory event that converted a putative RNA target into a 'locked' stable RNP. The ability to dimerize was also observed between members of the IMP family of zipcode-binding proteins, providing a multitude of further interaction possibilities within RNP granules and with the localization apparatus.
Figures





References
-
- Chartrand P., Singer,R.H. and Long,R.M. (2001) RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol., 17, 297–310. - PubMed
-
- Tekotte H. and Davis,I. (2002) Intracellular mRNA localization: motors move messages. Trends Genet., 18, 636–642. - PubMed
-
- Yaniv K. and Yisraeli,J.K. (2002) The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene, 287, 49–54. - PubMed
-
- Deshler J.O., Highett,M.I., Abramson,T. and Schnapp,B.J. (1998) A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol., 8, 489–496. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

