DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner
- PMID: 15316110
- PMCID: PMC520940
- DOI: 10.1105/tpc.104.022400
DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner
Abstract
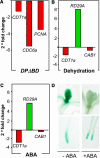
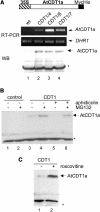
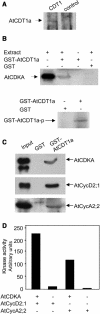
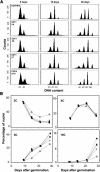
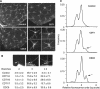
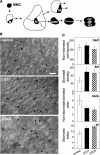
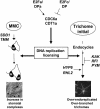
In eukaryotic cells, the function of DNA replication licensing components (Cdc6 and Cdt1, among others) is crucial for cell proliferation and genome stability. However, little is known about their role in whole organisms and whether licensing control interfaces with differentiation and developmental programs. Here, we study Arabidopsis thaliana CDT1, its regulation, and the consequences of overriding licensing control. The availability of AtCDT1 is strictly regulated at two levels: (1) at the transcription level, by E2F and growth-arresting signals, and (2) posttranscriptionally, by CDK phosphorylation, a step that is required for its proteasome-mediated degradation. We also show that CDC6 and CDT1 are key targets for the coordination of cell proliferation, differentiation, and development. Indeed, altered CDT1 or CDC6 levels have cell type-specific effects in developing Arabidopsis plants: in leaf cells competent to divide, cell proliferation is stimulated, whereas in cells programmed to undergo differentiation-associated endoreplication rounds, extra endocycles are triggered. Thus, we propose that DNA replication licensing control is critical for the proper maintenance of proliferative potential, developmental programs, and morphogenetic patterns.
Figures


Similar articles
-
Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation.Plant Cell. 2004 Sep;16(9):2350-63. doi: 10.1105/tpc.104.023978. Epub 2004 Aug 12. Plant Cell. 2004. PMID: 15308755 Free PMC article.
-
Expression and stability of Arabidopsis CDC6 are associated with endoreplication.Plant Cell. 2001 Dec;13(12):2671-86. doi: 10.1105/tpc.010329. Plant Cell. 2001. PMID: 11752380 Free PMC article.
-
The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis.Plant Cell. 2006 Sep;18(9):2224-35. doi: 10.1105/tpc.105.039651. Epub 2006 Aug 18. Plant Cell. 2006. PMID: 16920782 Free PMC article.
-
Down to the origin: Cdc6 protein and the competence to replicate.Trends Cell Biol. 2003 Mar;13(3):110-3. doi: 10.1016/s0962-8924(03)00024-2. Trends Cell Biol. 2003. PMID: 12628342 Review.
-
Regulation of DNA replication licensing.Curr Drug Targets. 2012 Dec;13(13):1588-92. doi: 10.2174/138945012803529965. Curr Drug Targets. 2012. PMID: 22998185 Review.
Cited by
-
Combined linkage and association mapping reveals CYCD5;1 as a quantitative trait gene for endoreduplication in Arabidopsis.Proc Natl Acad Sci U S A. 2012 Mar 20;109(12):4678-83. doi: 10.1073/pnas.1120811109. Epub 2012 Mar 5. Proc Natl Acad Sci U S A. 2012. PMID: 22392991 Free PMC article.
-
An RNA-seq transcriptome analysis of floral buds of an interspecific Brassica hybrid between B. carinata and B. napus.Plant Reprod. 2014 Dec;27(4):225-37. doi: 10.1007/s00497-014-0253-z. Epub 2014 Nov 15. Plant Reprod. 2014. PMID: 25398253
-
The plant cell cycle: Pre-Replication complex formation and controls.Genet Mol Biol. 2017;40(1 suppl 1):276-291. doi: 10.1590/1678-4685-GMB-2016-0118. Epub 2017 Mar 16. Genet Mol Biol. 2017. PMID: 28304073 Free PMC article.
-
Differentially methylated genes involved in reproduction and ploidy levels in recent diploidized and tetraploidized Eragrostis curvula genotypes.Plant Reprod. 2024 Jun;37(2):133-145. doi: 10.1007/s00497-023-00490-7. Epub 2023 Dec 6. Plant Reprod. 2024. PMID: 38055074 Free PMC article.
-
The MADS-box XAANTAL1 increases proliferation at the Arabidopsis root stem-cell niche and participates in transition to differentiation by regulating cell-cycle components.Ann Bot. 2016 Oct 1;118(4):787-796. doi: 10.1093/aob/mcw126. Ann Bot. 2016. PMID: 27474508 Free PMC article.
References
-
- Arentson, E., Faloon, P., Seo, J., Moon, E., Studts, J.M., Fremont, D.H., and Choi, K. (2002). Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene 21, 1150–1158. - PubMed
-
- Baulcombe, D.C., Saunders, G.R., Bevan, M.W., Mayo, M.A., and Harrison, B.D. (1986). Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321, 446–449.
-
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

