Control of plant trichome development by a cotton fiber MYB gene
- PMID: 15316114
- PMCID: PMC520936
- DOI: 10.1105/tpc.104.024844
Control of plant trichome development by a cotton fiber MYB gene
Abstract
Cotton (Gossypium spp) plants produce seed trichomes (cotton fibers) that are an important commodity worldwide; however, genes controlling cotton fiber development have not been characterized. In Arabidopsis thaliana the MYB gene GLABRA1 (GL1) is a central regulator of trichome development. Here, we show that promoter of a cotton fiber gene, RD22-like1 (RDL1), contains a homeodomain binding L1 box and a MYB binding motif that confer trichome-specific expression in Arabidopsis. A cotton MYB protein GaMYB2/Fiber Factor 1 transactivated the RDL1 promoter both in yeast and in planta. Real-time PCR and in situ analysis showed that GaMYB2 is predominantly expressed early in developing cotton fibers. After transferring into Arabidopsis, GL1::GaMYB2 rescued trichome formation of a gl1 mutant, and interestingly, 35S::GaMYB2 induced seed-trichome production. We further demonstrate that the first intron of both GL1 and GaMYB2 plays a role in patterning trichomes: it acts as an enhancer in trichome and a repressor in nontrichome cells, generating a trichome-specific pattern of MYB gene expression. Disruption of a MYB motif conserved in intron 1 of GL1, WEREWOLF, and GaMYB2 genes affected trichome production. These results suggest that cotton and Arabidopsis use similar transcription factors for regulating trichomes and that GaMYB2 may be a key regulator of cotton fiber development.
Figures
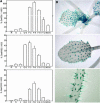

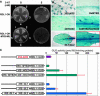


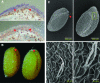
References
-
- Abe, M., Takahashi, T., and Komeda, Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494. - PubMed
-
- Cai, X.L., Wang, Z.Y., Xing, Y.Y., Zhang, J.L., and Hong, M.M. (1998). Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. - PubMed
-
- Callis, J., Fromm, M., and Walbot, V. (1987). Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200. - PubMed
-
- Cedroni, M.L., Cronn, R.C., Adams, K.L., Wilkins, T.A., and Wendel, J.F. (2003). Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol. Biol. 51, 313–325. - PubMed
-
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

