Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation
- PMID: 15317759
- PMCID: PMC516813
- DOI: 10.1128/JB.186.17.5557-5566.2004
Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation
Abstract
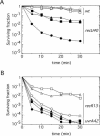
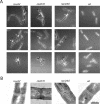
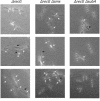
Bacillus subtilis mutants classified within the epsilon (ruvA, DeltaruvB, DeltarecU, and recD) and eta (DeltarecG) epistatic groups, in an otherwise rec+ background, render cells impaired in chromosomal segregation. A less-pronounced segregation defect in DeltarecA and Deltasms (DeltaradA) cells was observed. The repair deficiency of addAB, DeltarecO, DeltarecR, recH, DeltarecS, and DeltasubA cells did not correlate with a chromosomal segregation defect. The sensitivity of epsilon epistatic group mutants to DNA-damaging agents correlates with ongoing DNA replication at the time of exposure to the agents. The Deltasms (DeltaradA) and DeltasubA mutations partially suppress the DNA repair defect in ruvA and recD cells and the segregation defect in ruvA and DeltarecG cells. The Deltasms (DeltaradA) and DeltasubA mutations partially suppress the DNA repair defect of DeltarecU cells but do not suppress the segregation defect in these cells. The DeltarecA mutation suppresses the segregation defect but does not suppress the DNA repair defect in DeltarecU cells. These results result suggest that (i) the RuvAB and RecG branch migrating DNA helicases, the RecU Holliday junction (HJ) resolvase, and RecD bias HJ resolution towards noncrossovers and that (ii) Sms (RadA) and SubA proteins might play a role in the stabilization and or processing of HJ intermediates.
Figures


References
-
- Alonso, J. C., A. C. Stiege, and G. Luder. 1993. Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH, and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 239:129-136. - PubMed
-
- Ayora, S., F. Rojo, N. Ogasawara, S. Nakai, and J. C. Alonso. 1996. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 256:301-318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

