Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain
- PMID: 15317764
- PMCID: PMC516811
- DOI: 10.1128/JB.186.17.5603-5613.2004
Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain
Abstract
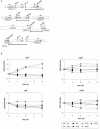
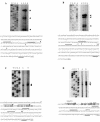
The cold shock proteins are small peptides that share a conserved domain, called the cold shock domain (CSD), that is important for nucleic acid binding. The Caulobacter crescentus genome has four csp genes that encode proteins containing CSDs. Three of these (cspA, cspB, and cspC) encode peptides of about 7 kDa and are very similar to the cold shock proteins of other bacteria. Analysis by reverse transcription-PCR of the fourth gene (cspD), which was previously annotated as encoding a 7-kDa protein, revealed that the mRNA is larger and probably encodes a putative 21-kDa protein, containing two CSDs. A search in protein sequences databases revealed that this new domain arrangement has thus far only been found among deduced peptides of alpha-proteobacteria. Expression of each Caulobacter csp gene was studied both in response to cold shock and to growth phase, and we have found that only cspA and cspB are induced by cold shock, whereas cspC and cspD are induced at stationary phase, with different induction rates. The transcription start sites were determined for each gene, and a deletion mapping of the cspD promoter region defined a sequence required for maximal levels of expression, indicating that regulation of this gene occurs at the transcriptional level. Deletion of cspA, but not cspD, caused a reduction in viability when cells were incubated at 10 degrees C for prolonged times, suggesting that cspA is important for adaptation to a low temperature.
Figures





Similar articles
-
Cold shock genes cspA and cspB from Caulobacter crescentus are posttranscriptionally regulated and important for cold adaptation.J Bacteriol. 2012 Dec;194(23):6507-17. doi: 10.1128/JB.01422-12. Epub 2012 Sep 21. J Bacteriol. 2012. PMID: 23002229 Free PMC article.
-
CspC and CspD are essential for Caulobacter crescentus stationary phase survival.Arch Microbiol. 2010 Sep;192(9):747-58. doi: 10.1007/s00203-010-0602-8. Epub 2010 Jul 6. Arch Microbiol. 2010. PMID: 20607520
-
Identification and regulation of cold-inducible factors of Bordetella bronchiseptica.Microbiology (Reading). 2005 Jun;151(Pt 6):1895-1909. doi: 10.1099/mic.0.27785-0. Microbiology (Reading). 2005. PMID: 15941997
-
The cold-shock response--a hot topic.Mol Microbiol. 1994 Mar;11(5):811-8. doi: 10.1111/j.1365-2958.1994.tb00359.x. Mol Microbiol. 1994. PMID: 8022259 Review.
-
The CspA family in Escherichia coli: multiple gene duplication for stress adaptation.Mol Microbiol. 1998 Jan;27(2):247-55. doi: 10.1046/j.1365-2958.1998.00683.x. Mol Microbiol. 1998. PMID: 9484881 Review.
Cited by
-
Transcriptomic analysis of the stationary phase response regulator SpdR in Caulobacter crescentus.BMC Microbiol. 2016 Apr 12;16:66. doi: 10.1186/s12866-016-0682-y. BMC Microbiol. 2016. PMID: 27072651 Free PMC article.
-
Extended -10 motif is critical for activity of the cspA promoter but does not contribute to low-temperature transcription.J Bacteriol. 2005 Sep;187(18):6584-9. doi: 10.1128/JB.187.18.6584-6589.2005. J Bacteriol. 2005. PMID: 16159795 Free PMC article.
-
Cold shock genes cspA and cspB from Caulobacter crescentus are posttranscriptionally regulated and important for cold adaptation.J Bacteriol. 2012 Dec;194(23):6507-17. doi: 10.1128/JB.01422-12. Epub 2012 Sep 21. J Bacteriol. 2012. PMID: 23002229 Free PMC article.
-
Can Halophilic and Psychrophilic Microorganisms Modify the Freezing/Melting Curve of Cold Salty Solutions? Implications for Mars Habitability.Astrobiology. 2020 Sep;20(9):1067-1075. doi: 10.1089/ast.2019.2094. Epub 2020 Aug 20. Astrobiology. 2020. PMID: 32833498 Free PMC article.
-
Growth-dependent activity of the cold shock cspA promoter + 5' UTR and production of the protein CspA in Staphylococcus aureus Newman.BMC Res Notes. 2017 Jun 27;10(1):232. doi: 10.1186/s13104-017-2557-1. BMC Res Notes. 2017. PMID: 28655334 Free PMC article.
References
-
- Abyzov, S. S., I. N. Mitskevich, M. N. Poglazova, N. I. Barkov, V. Y. Lipenkov, N. E. Bobin, B. B. Koudryashov, V. M. Pashkevich, and M. V. Ivanov. 2001. Microflora in the basal strata at Antarctic ice core above the Vostok lake. Adv. Space Res. 28:701-706. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 27:403-410. - PubMed
-
- Bouvet, P., and A. P. Wolffe. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77:931-941. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

