Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly
- PMID: 15317787
- PMCID: PMC516821
- DOI: 10.1128/JB.186.17.5819-5825.2004
Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly
Abstract
The ribosomal protein L11 in bacteria is posttranslationally trimethylated at multiple amino acid positions by the L11 methyltransferase PrmA, the product of the prmA gene. The role of L11 methylation in ribosome function or assembly has yet to be determined, although the deletion of Escherichia coli prmA has no apparent phenotype. We have constructed a mutant of the extreme thermophile Thermus thermophilus in which the prmA gene has been disrupted with the htk gene encoding a heat-stable kanamycin adenyltransferase. This mutant shows no growth defects, indicating that T. thermophilus PrmA, like its E. coli homolog, is dispensable. Ribosomes prepared from this mutant contain unmethylated L11, as determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), and are effective substrates for in vitro methylation by cloned and purified T. thermophilus PrmA. MALDI-TOF MS also revealed that T. thermophilus L11 contains a total of 12 methyl groups, in contrast to the 9 methyl groups found in E. coli L11. Finally, we found that, as with the E. coli methyltransferase, the ribosomal protein L11 dissociated from ribosomes is a more efficient substrate for in vitro methylation by PrmA than intact 70S ribosomes, suggesting that methylation in vivo occurs on free L11 prior to its incorporation into ribosomes.
Figures
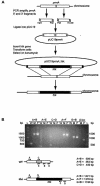
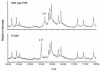
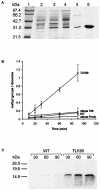
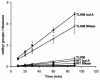
Similar articles
-
Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA.EMBO J. 2007 Jan 24;26(2):567-77. doi: 10.1038/sj.emboj.7601508. Epub 2007 Jan 11. EMBO J. 2007. PMID: 17215866 Free PMC article.
-
Ribosomal protein methylation in Escherichia coli: the gene prmA, encoding the ribosomal protein L11 methyltransferase, is dispensable.Mol Microbiol. 1994 Dec;14(5):947-58. doi: 10.1111/j.1365-2958.1994.tb01330.x. Mol Microbiol. 1994. PMID: 7715456
-
Crystallization and preliminary X-ray diffraction analysis of ribosomal protein L11 methyltransferase from Thermus thermophilus HB8.Acta Crystallogr D Biol Crystallogr. 2003 May;59(Pt 5):930-2. doi: 10.1107/s0907444903004554. Epub 2003 Apr 25. Acta Crystallogr D Biol Crystallogr. 2003. PMID: 12777815
-
Ribosomal proteins from Thermus thermophilus for structural investigations.Biochimie. 1992 Apr;74(4):327-36. doi: 10.1016/0300-9084(92)90110-z. Biochimie. 1992. PMID: 1637860 Review.
-
Confounders of mutation-rate estimators: selection and phenotypic lag in Thermus thermophilus.Mutat Res. 2013 Sep;749(1-2):16-20. doi: 10.1016/j.mrfmmm.2013.07.006. Epub 2013 Aug 2. Mutat Res. 2013. PMID: 23916418 Free PMC article. Review.
Cited by
-
aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism.Mol Cell Proteomics. 2016 Sep;15(9):2908-23. doi: 10.1074/mcp.M115.057778. Epub 2016 Jun 21. Mol Cell Proteomics. 2016. PMID: 27329856 Free PMC article.
-
The functional diversity of protein lysine methylation.Mol Syst Biol. 2014 Apr 8;10(4):724. doi: 10.1002/msb.134974. Mol Syst Biol. 2014. PMID: 24714364 Free PMC article. Review.
-
Developing limited proteolysis and mass spectrometry for the characterization of ribosome topography.J Am Soc Mass Spectrom. 2007 Jul;18(7):1304-17. doi: 10.1016/j.jasms.2007.03.028. Epub 2007 Apr 6. J Am Soc Mass Spectrom. 2007. PMID: 17521915 Free PMC article.
-
Identification of Genes Involved in Antifungal Activity of Burkholderia seminalis Against Rhizoctonia solani Using Tn5 Transposon Mutation Method.Pathogens. 2020 Sep 27;9(10):797. doi: 10.3390/pathogens9100797. Pathogens. 2020. PMID: 32992669 Free PMC article.
-
Ribosome engineering to promote new crystal forms.Acta Crystallogr D Biol Crystallogr. 2012 May;68(Pt 5):578-83. doi: 10.1107/S0907444912006348. Epub 2012 Apr 17. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22525755 Free PMC article.
References
-
- Agrawal, R. K., J. Linde, J. Sengupta, K. H. Nierhaus, and J. Frank. 2001. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 311:777-787. - PubMed
-
- Alix, J. H. 1992. Extrinsic factors in ribosome assembly, p. 173-184. In K. H. Nierhaus, A. R. Subramanian, V. A. Erdmann, F. Franceschi, and B. Wittmann-Liebold (ed.), The translational apparatus. Plenum Publishing, New York, N.Y.
-
- Allen, P. N., and H. F. Noller. 1991. A single base substitution in 16S ribosomal RNA suppresses streptomycin dependence and increases the frequency of translational errors. Cell 66:141-148. - PubMed
-
- Bujnicki, J. M. 2000. Sequence, structural, and evolutionary analysis of prokaryotic ribosomal protein L11 methyltransferases. Acta Microbiol. Pol. 49:19-28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

