A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall
- PMID: 15317792
- PMCID: PMC516832
- DOI: 10.1128/JB.186.17.5865-5875.2004
A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall
Abstract
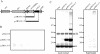
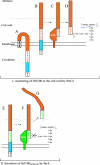
The important human pathogen Streptococcus pyogenes (group A streptococcus GAS), requires several surface proteins to interact with its human host. Many of these are covalently linked by a sortase enzyme to the cell wall via a C-terminal LPXTG motif. This motif is followed by a hydrophobic region and charged C terminus, which are thought to retard the protein in the cell membrane to facilitate recognition by the membrane-localized sortase. Previously, we identified two sortase enzymes in GAS. SrtA is found in all GAS strains and anchors most proteins containing LPXTG, while SrtB is present only in some strains and anchors a subset of LPXTG-containing proteins. We now report the presence of a third sortase in most strains of GAS, SrtC. We show that SrtC mediates attachment of a protein with a QVPTGV motif preceding a hydrophobic region and charged tail. We also demonstrate that the QVPTGV sequence is a substrate for anchoring of this protein by SrtC. Furthermore, replacing this motif with LPSTGE, found in the SrtA-anchored M protein of GAS, leads to SrtA-dependent secretion of the protein but does not lead to its anchoring by SrtA. We conclude that srtC encodes a novel sortase that anchors a protein containing a QVPTGV motif to the surface of GAS.
Figures




References
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. - PMC - PubMed
-
- Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, P. F. Garcia-del, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

