Characterization of Salmonella enterica subspecies I genovars by use of microarrays
- PMID: 15317794
- PMCID: PMC516822
- DOI: 10.1128/JB.186.17.5883-5898.2004
Characterization of Salmonella enterica subspecies I genovars by use of microarrays
Abstract
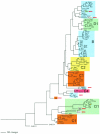
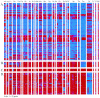
Subspecies 1 of Salmonella enterica is responsible for almost all Salmonella infections of warm-blooded animals. Within subspecies 1 there are over 2,300 known serovars that differ in their prevalence and the diseases that they cause in different hosts. Only a few of these serovars are responsible for most Salmonella infections in humans and domestic animals. The gene contents of 79 strains from the most prevalent serovars were profiled by microarray analysis. Strains within the same serovar often differed by the presence and absence of hundreds of genes. Gene contents sometimes differed more within a serovar than between serovars. Groups of strains that share a distinct profile of gene content can be referred to as "genovars" to distinguish them from serovars. Several misassignments within the Salmonella reference B collection were detected by genovar typing and were subsequently confirmed serologically. Just as serology has proved useful for understanding the host range and pathogenic manifestations of Salmonella, genovars are likely to further define previously unrecognized specific features of Salmonella infections.
Figures

References
-
- Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. - PubMed
-
- Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

